Artigo
| Biomolecules produced in liquid-state fermentation by a marine-derived fungus, Penicillium roqueforti |
|
Roberto MiosoI,*; Francisco J. T. MaranteI; Irma H. Bravo de LagunaII; Juan E. G. GonzálezIII; Juan J. S. RodríguezIII
IDepartamento de Química, Universidad de Las Palmas de Gran Canaria, Gran Canaria 35017, Spain Recebido em 04/06/2013 *e-mail: robertomioso@yahoo.co.uk Screening of biomass of a new marine-derived strain of Penicillium roqueforti, as produced by liquid-state fermentation, led to the identification of several volatile organic compounds active in the fatty acid pathway as well as fragments produced by their catabolism, terpenoids, and metabolites from the shikimic acid pathway. In addition, five non-volatile organic compounds, triolein, ergosterol peroxide, 9(11)-dehydroergosterol peroxide, 4-hydroxybenzaldehyde, and d-mannitol, were isolated and identified by spectroscopy. The results showed that this fungal strain did not produce any mycotoxin in the culture conditions applied, and thus is useful for industrial applications, where high value-added biomolecules are generated. INTRODUCTION The filamentous fungus Penicillium roqueforti is well-known for its use in biotechnological applications and has been extensively used in the dairy industry to add flavor and veining to internally mould-ripened blue cheeses.1 It is a common contaminant mould found in silages, food, and feed, and its ability to produce a large number of biologically active extrolites, including various toxins, has attracted the interest of many researchers.2 Although it is described as a terrestrial fungus, some studies have shown that P. roqueforti strains have high salt tolerance,3 with spore germination inhibited only at sodium chloride concentrations of over 100 g L-1.4 This extraordinary tolerance to salt, along with its tolerance to high osmotic pressure, indicates that this hyphomycete adapts to life as a marine facultative fungus.5 Note that the average seawater salinity is 35 g L-1. Moreover, under certain culture conditions, this fungus has shown its pronounced capacity to biosynthesize secondary metabolites,6 some of which have antiparasitic7 and bacteriostatic properties.8 Its proteolytic enzymes allow it to be applied in various processes, such as removal of scales of some fish, in the fishing industry9 and in biotechnology for the production of high-quality fat from food waste.10 With regard to the P. roqueforti chemical composition, it is worth noting the extensive research performed on its fatty acid profile and lipid metabolism.11 Chromatographic studies on the total lipid fraction of P. roqueforti have shown the presence of palmitic, stearic, oleic, and linoleic acids sterified in the form of acyl-glycerides and free fatty acids, whereas the more polar lipids are composed of phospholipids and glycolipids,12 and the free steroids have an ergosterol skeleton.13 Further data are available on the biogenesis of unsaturated fatty acids depending on the phases of growth.14 However, the applicability of this fungi in foodstuffs became limited after the discovery that these organisms are also capable of producing dangerous secondary metabolites.15,16 These toxic strains can be identified using GC-MS in line with the chemical tracers proposed by Demyttenaere et al.,17 Jelen,18 and Calvert et al.19 P. roqueforti produces several mycotoxins, such as PR-toxin, roquefortine C, mycophenolic acid, patulin, and penicillic acid,20 some of which are known to be generally unstable or incapable of causing serious damage at low concentrations.21 Although they can occur under natural conditions in feed, both roquefortine C and mycophenolic acid are considered to be low-toxicity mycotoxins with less significance.20-22 PR-toxin is the most significant because it is reported to cause damage to the liver and kidney in rats16 and is also potentially carcinogenic.23 However, it should be noted that fungal metabolite production depends on several variables such as the isolate,24 growth medium, and environmental factors.25 Thus, the present study examined the metabolites biosynthesized by the fungus P. roqueforti that was isolated from marine biota and grown in marine culture media. Detailed screening of mycelia was performed and volatile and non-volatile organic compounds were identified using GC-MS and NMR spectroscopy. It is anticipated that the chemo-specific information may offer crucial information for improving its biotechnological applications.
EXPERIMENTAL Isolation and identification of the fungus The halotolerant fungal strain was obtained from marine water collected at the intertidal zone of the "La Laja" beach, Gran Canaria, the Canary Islands, Spain. Water samples were randomly collected in sterile flasks (24 units; 125 mL each) at 0.2-0.5 m depth in accordance with the procedures proposed by Seymour and Fuller.26 Each water sample (0.5 mL, undiluted) was spread on Petri dishes, and the plates were incubated at 26 ºC (±2 ºC) for 21 days. The isolation process and strain purification were performed on Petri dishes using a modified KMV solid medium containing 1 g yeast extract, 1 g hydrolyzed gelatin, 1 g peptone, 5 g glucose, and 12 g bacteriological agar in 1 L filtered seawater (salinity, 35 ppt). The fungus was identified as P. roqueforti using morphological criteria defined by CABI Bioscience, Surrey, UK (see supplementary material), and a voucher specimen was deposited at the laboratory for future references under the accession number PA 002. Liquid-state fermentation After the purification of colonies, the mycelial spores were scraped and transferred to Erlenmeyer flasks (2 L capacity) containing the sterile modified KMV broth, which was autoclaved at 121 ºC for 20 min. The modified KMV broth contained 1 g yeast extract, 1 g hydrolyzed gelatin, 1 g peptone, and 5 g glucose in 1 L filtered seawater. Fungal biomass production was performed in liquid-state fermentation in static polypropylene boxes (83 x 46 x 18 cm) that were previously sterilized with sodium hypochlorite and steamed for 5 min. Once the steam was condensed, water was drained out of the boxes. The spore solution was then homogenized for 10 min in a magnetic mixer using a bar stir and transferred directly to each box (20 units with 1.2 L of culture broth/unit). After 10-12 days of incubation at room temperature (22-25 ºC), the supernatant mycelia were separated by filtration and dried by IR radiation. Apparatus and analytical methods Normal-phase chromatography was performed on silica gel (Scharlau) using a 0.06-0.2 mm particle sized adsorbent and a 0.04-0.06 mm particle sized stationary phase. Chromatography was performed at medium (Büchi Chromatography System) or low pressure using Fluid Metering Inc. motors connected in series with an Ace Glass Inc. column. Reverse-phase chromatography was performed on a LiChroprep RP-18 (40-63 µm particle size; Merck) column connected to a low-pressure chromatography system also based on the Fluid Metering Inc. apparatus. Size-exclusion chromatography was performed on lipophilic Sephadexr LH-20 (Sigma). The column was eluted first with anhydrous methanol (2 h) and then with a mixture of CH2Cl2/CH3OH (50:50,2 h). The extracts were applied at the top of the column and eluted with CH2Cl2/CH3OH (50:50) at a rate of 1.0 mL min-1. Normal-phase TLC was performed on silica gel plates (0.25 mm diameter; Tracer Analitica) using a combination of n-hexane, ethyl acetate, chloroform, and methanol as the eluent, in the proportions specified for each. Reverse-phase TLC was performed on RP-18F254 plates (0.25 mm; Merck) using CH3CN/CH3OH/H2O (80:18:2) as the other mobile phase. In all cases, spots were revealed by spraying with oleum [sulfuric acid (4%) + acetic acid (80%) + water (16%)] and heating at 120 ºC for 20 min. Normal-phase semipreparative HPLC was performed using an Alltech Econosphere silica column (10 µm particle size, 250 x 4.6 mm, and 100 Å pore size), and reverse-phase semipreparative HPLC was performed on a Waters ODS column (10 µm particle size, 250 x 4.6 mm, and 100 Å pore size). Both these processes were conducted using a semipreparative HPLC apparatus coupled with a Spectra-physics P100 isocratic pump and in line with a Hewlett Packard 1050 UV-vis variable wavelength detector working at room temperature (26 ºC). Analytical chromatography was performed using a Shimadzu HPLC system with an LC-9A pump connected to a UV SPD-6AV detector (254 nm). The conditions used for the normal-phase column were combinations of n-hexane and ethyl acetate as the eluent; in case of the size-exclusion chromatography column (Shodex OH Pak SB 806 HQ), a mixture of water and 0.05% sodium azide was used as the eluent. An eluent flow rate of 1.0 mL min-1 was used for all analyses. Infrared spectra were recorded on a Shimadzu FTIR-8400S spectrophotometer, with chloroform (Merck) as a solvent for spectroscopy. The samples were sandwiched between two sodium chloride plates, and the spectrum was calibrated against the 1603 cm-1 band of polystyrene. 1H, 13C, and 2D-NMR experiments were recorded at 250 or 300 MHz on AC or AMX Bruker apparatus, respectively. A Varian UNITY INOVA 400 MHz NMR spectrometer was used for high-resolution analysis. Tetramethylsilane was used as an internal standard for 1H, and deuterated chloroform (δ 77.00) or deuterated methanol (δ 49.00) was used for the calibration of 13-carbon NMR spectra. Electrospray ionization mass spectrometry was performed either at low or high resolution with a common electron impact mass spectrometer (IE) or by fast atom bombardment (FAB). Positive mode was performed on a FAB-MS at 70 eV with a FISONS VG Micromass Autospec apparatus, with NBA (3-nitrobenzyl alcohol) as the matrix. Melting points were established using a Gallenkamp apparatus and were left uncorrected. Gas chromatography-mass spectrometry (GC-MS) was performed on a chromatograph model Varian CP3800 with an ion-trap mass spectrometer model Saturn 2000 and under the following conditions: CP-Sil 8 low bleed/MS capillary column. The injector temperature was kept isothermal at 270 ºC, initial split conditions on, and 0.01 min off and 5 min on, with a split ratio of 1:50; the oven was set at 50 ºC for 5 min and then ramped at 15 ºC min-1 to 250 ºC and held for 10 min (for a total run time of 28.33 min for each sample) with a flux of 1 mL min-1, using the mass detector in the EI mode (20-400 m/z). Compounds lacking reference standard were quantified using the response factor for alkanes (Dr. Ehrenstorfer GmbH Alkanes-Mix 10), fatty acid methyl esters (SupelcoTM 37 Component FAME Mix), 1-alkenes (Fluka Chemika), and 1-alkanols (Fluka Chemika). The remaining compounds were assigned by structural analogy to the above. Thus, tetradecanoic acid 1-(hydroxymethyl)-1,2-ethanediyl ester was assigned to the factor obtained experimentally for hexadecanoic acid methyl ester (764.117 x 10-12 mg K counts-1). This same factor was used for other FAME such as tetra-unsaturated 6,9,12,15-docosatetraenoic acid methyl ester.
RESULTS AND DISCUSSION Chemical analysis Each box of culture broth (1.2 L) yielded 38.71-49.67 g of fresh mycelia, corresponding to 3.75-5.42 g of dry matter. The total wet mass of mycelia resulting from the sum of yields was 888.34 g, which produced 89.9 g of dry matter after IR desiccation. The crude extract was obtained by maceration in CH2Cl2 (x3,24 h) and CH3OH (x3,24 h) at room temperature. After filtration, evaporation, and vacuum desiccation, 14.8 g of brown oil was obtained. The whole extract was fractionated by polarity in a liquid-liquid extraction system, according to a modified version of the Kupchan method.27 The process flow diagram can be found in supplementary material (Figure 2S). Each fraction was screened using chromatography (column chromatography, size-exclusion chromatography, and thin-layer chromatography) and analyzed using GC-MS (for volatile compounds) or spectroscopy (NMR, MS, and IR), allowing the identification of the following categories of compounds: Volatile organic compounds Volatile organic compounds, i.e., compounds that volatilize in gas chromatograph injector at temperatures of 270 ºC, were identified by GC-MS (Table 1) and classified by structural criteria (Figures 1-3) as follows: n-alkanes (1); 1-alkenes (2); 1-alkanols (3); 2-alkyl-1-alkanols (4); saturated (5) and unsaturated (7) free fatty acids; fatty acid amides (10); saturated (6) and unsaturated (8,11-14) fatty acid methyl and ethyl esters; unsaturated triglycerides (15) and diglycerides (16); unsaturated monoglycerides (17); wax esters (19 and 20); lipid catabolites (21 and 22); mono-, sesqui-, and straight-chain terpenes (23-25); and an aromatic hydrocarbon from the shikimic acid route (26).
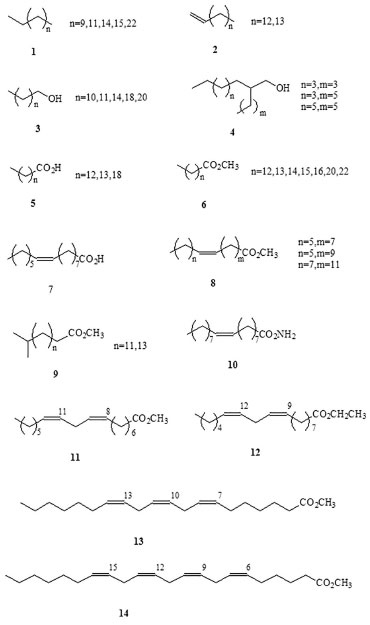 Figure 1. Volatile lipid compounds identified in Penicillium roqueforti
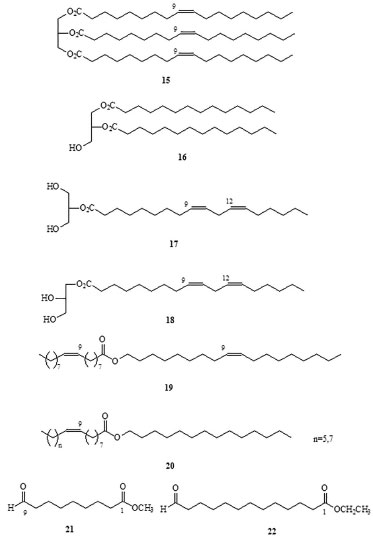 Figure 2. Volatile lipid compounds identified in Penicillium roqueforti
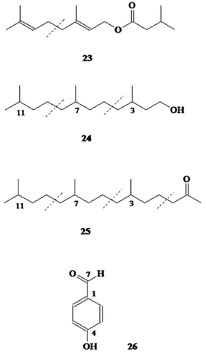 Figure 3. Volatile terpenoid/shikimate compounds identified in Penicillium roqueforti
Occurrence of methyl-branched fatty acids Methyl-branched fatty acids are present as minor lipid components in various living organisms and as major components of lipids in various bacteria.28 It has been ascertained that they are formed by the selective incorporation of methylmalonyl-CoA, catalyzed by the fatty acid synthetase enzyme.29 This biogenetic pathway is a characteristic of bacteria that produce relatively high concentrations of these iso-methyl-branched fatty acids, which are therefore accepted as molecular markers of organic matter produced by these organisms.30 Therefore, identification of methyl ester in 14-methyl-pentadecanoic acid (9, n = 11) and 16-methyl-heptadecanoic acid (9, n = 13), the two iso-methyl-branched fatty acids derivatives, is an indirect evidence of the presence of bacteria associated with this fungus. Apart from the GC-MS fingerprint, the iso-methyl-substitution proposed in 9 (n = 11 and 13) was confirmed by the relatively intense fragment ion peak at M+-43 (m/z = 227 and 255, respectively) observed using GC-MS, together with a decrease in the intensity of the M+-29 (m/z = 241 and 269, respectively) fragment.31 Non-volatile organic compounds Non-volatile compounds obtained from P. roqueforti mycelia were fractionated and measured by gravimetric analysis and integration of the 1H-NMR spectrum and were classified as high-molecular weight alkanes (1.29%), waxes (0.37%), unsaturated steroidal waxes (1.11%), other waxes (2.73%), unsaturated triglycerides [triolein (15) and other, 2.56%], ergosterol peroxide (27, 12.63%), 9(11)-dehydroergosterol peroxide (DHEP; 28, 2.15%), 4-hydroxybenzaldehyde (26, 0.92%), unidentified phospho- and glycolipids (0.31%), mannitol (29, 5.0%), and unidentified polyhydroxy compounds (70.93%). Note that some of these described compounds were also detected as volatile components in the previous section. The major substances, triolein, ergosterol peroxide (EP), 9(11)-DHEP, and mannitol, were the only compounds to be purified and characterized in these non-volatile lipid compounds (Figure 4). Among monocyclic aromatic compounds from the shikimic acid route, 4-hydroxybenzaldehyde could be purified and characterized (Figure 3).
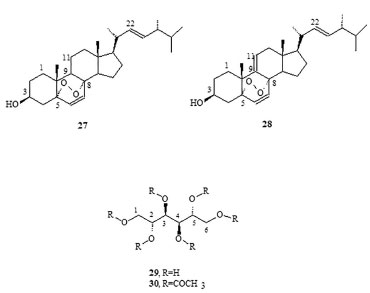 Figure 4. Non-volatile compounds identified in Penicillium roqueforti
Triolein (15) was obtained from one of the less polar fractions of the mycelia extract as yellowish oil and was identified using IR, 1H-NMR, 13C-NMR, 1H-1H-(COSY, TOCSY, NOESY), DEPT_135, HSQC, HMBC, and mass spectroscopic data. Although several factors may affect either the fatty acid composition or the percentage of total lipids found in fungi,32 fatty acid analysis of the cultured P. roqueforti indicates a limited capacity of lipid accumulation in this strain. However, the characteristic oleic acid as a major constituent of the unsaturated fatty acid fraction (0.4% DW of mycelia) offers functional benefits of oxidative stability and nutritional attributes to this fungus. Another biomolecule that was isolated in its purest form was 5α, 8α-epidioxyergosta-6,22-dien-3β-ol (27) usually denominated ergosterol peroxide (EP). The product, which crystallizes from methanol, has a melting point of 178-180 ºC and was identified using IR, 1H-NMR, 13C-NMR, 1H-1H-(COSY, TOCSY, NOESY), DEPT_135, HSQC, HMBC, and mass spectroscopic data (Table 2).

Previous studies have offered contradictory explanations for the origin of sterol peroxides that may be formed by the photo-oxygenation of sterols with double conjugated bonds at C-5/C-6 carbons and at C-7/C-8 carbons.33 Thus, EP in extracts from fungi was regarded as an artifact rather than as a natural product, which was produced by the photo-oxygenation of ergosterol during the experimental procedures or was possibly sensitized by fungal pigments during mycelia growth.34 Although there are no references to the existence of sterol peroxides as biological membrane constituents, enzymatic and photo-oxidative conversions of ergosterol into EP were observed in vivo in Penicillium sp. and Gibberella sp.22 Similarly, Sheikh and Djerassi35 working with a sterol mixture from the marine sponge Tethya aurantia suggested that EP was formed by biological processes. EP is also a major antitumor sterol produced by edible or medicinal mushrooms.36-38 This compound can be either extracted from another filamentous fungal species such as Paecilomyces variotii, 39 P. tenuipes, 40 and P. herquei41 or synthesized from ergosterol by photosensitized oxygenation with eosin.42 The 9(11)-dehydro derivative (28) was not isolated in its purest form, and it was found with the aforementioned EP (27) in the form of a white solid, with a melting point (Mp) of 171 ºC-176 ºC [α]D20 = -9.7 (CHCl3, c 1.2), and was identified using IR, 1H-NMR, 13C-NMR, 1H-1H-(COSY, TOCSY, NOESY), DEPT_135, HSQC, HMBC, and mass spectroscopic data. The 1H-NMR spectrum showed two superimposed AB systems centered at δ 6.41 and 6.48, respectively, with J = 8.7 Hz for both. After consulting the literature, this material was recognized as a mixture of EP and its 9(11)-dehydro derivative.43 The quantitative ratio of these products in the mixture was obtained by integrating the olefinic protons in the AB system NMR spectrum (400 MHz), resulting in a mix of EP (79%) and 9(11)-DHEP (21%), structures 27 and 28, respectively. This quantitative relation was confirmed using the optical rotation method. A graph was then drawn using the experimental values of optical rotation of the mixture versus the percentage of EP. The graph of the optical rotation data provided by Fisch et al.42 and Mediavilla43 (Table 1S in the supplementary material) showed a linear relationship between the value of [α]D and the percentage of EP, where % EP = 70 - 0.91 x [α]D By introducing the value [α]D20 = -9.7 into this equation, the optical rotation obtained for this material was 78.8% for EP, which was consistent with the 79.0% optical rotation obtained by integrating the 1H-NMR spectrum. Finally, it should be noted that in the 13C-NMR spectrum of this mixture, six small signs of olefinic carbons can be detected from the DHEP, with the signals assigned relevant for C-9 carbon at δ 130.970 (δ 51.081 in EP) and C-11 carbon at δ 125.00 (δ 20.866 in EP). Moreover, the 1H-NMR spectrum of this mixture gives a signal at δ 5.48 (0.21 H, m) that has been assigned to vinyl proton on C-11 carbon. Another non-volatile compound, 4-hydroxybenzaldehyde (26), was obtained pure by crystallization of methanol (Mp = 121-122 ºC). Its structure was elucidated from its spectroscopic data, mainly NMR spectroscopy (Table 3).
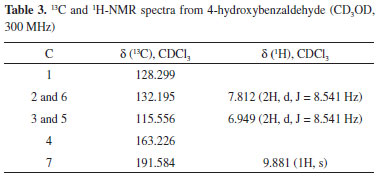
Although it is the first time that this has been described in fungi, this substance is structurally related to other previously known metabolites in the said organisms that are involved in the shikimic acid pathway, such as oxime-2-(4-hydroxyphenyl)-2-oxo acetaldehyde, a metabolite previously isolated from P. olsonii.44 Finally, d-mannitol, the last non-volatile compound, was obtained from the most polar fraction, and was characterized based on the spectroscopic data of its hexa-acetate derivative. The product was crystallized in the shape of transparent hexagonal crystals. The melting point of the mix (with authentic mannitol acetate) was 123 ºC-124 ºC and the optical rotation was [α]D20 = +23.68 (CHCl3, c 1.93), which are consistent with the literature.45 Polyhydroxy alcohols or sugar alcohols are biomolecules produced by many organisms, including plants, bacteria, and fungi.46 In fungi, mannitol is the most common polyol found in large quantities in spores, fruiting bodies, sclerotia, and mycelia.47 In Aspergilus niger conidiophores, for example, this compound may make up 10%-15% of the dry weight.48 Thus, the use of raw materials derived from renewable sources remains an excellent choice for the development of new high value-added substances. Mannitol is produced commercially by catalytic hydrogenation of fructose syrups or by inverting sugar with the co-production of another sugar alcohol sorbitol. Typically, hydrogenation of a 50/50 fructose/glucose mixture results in a 30/70 mixture of mannitol and sorbitol.49 For better yield, some alternative processes based on the use of microbes have been suggested in the literature. For example, yeast, fungi, and lactic acid bacteria in particular are known to produce mannitol effectively without the co-formation of sorbitol.50 Thus, the production of mannitol by fermentation from alternative sources such as the filamentous fungi P. roqueforti could have interesting applications because mannitol has widespread use in clinical medicine,51 with applications in the food and cosmetics industries.52 Absence of mycotoxins Strain development of filamentous fungi has focused both on productivity and safety, and the latter is widely exploited as factories of cells in the food and beverage industry worldwide.53 In some cases, related strains may produce toxins because of which Penicillium mycotoxins have been well documented.54 However, because of the use of the present methodology for biomass production, it was not possible to detect potential bioactive toxic compounds in the mycelia of P. roqueforti nor in the intermediary metabolites involved in a biogenetic route that could produce these substances. Consistent with this observation, no non-volatile organic compound was found that could bind biogenetically with the mycotoxins such as patulin, PR toxin, and mycophenolic acid54,55 that have been previously described in the literature for Penicillium sp. Moreover, there are no indications that the studied strain may produce botryodiplodin, a mycotoxin described by Moreau et al.56 in a P. roqueforti strain that did not produce the PR-toxin. However, such volatile compounds were found in the NIST and Wiley mass spectral database using the Varian Saturn GC-MS equipment. This suggests the potential use of this strain in food (both animal and human).
CONCLUSIONS In all, 50 volatile compounds were identified in the P. roqueforti mycelia, which included n-alkanes; 1-alkenes; 1-alkanols; 2-alkyl-1-alkanols; saturated and unsaturated free fatty acids; fatty acid amides; saturated and unsaturated fatty acid methyl and ethyl esters; unsaturated triglycerides and diglycerides; unsaturated monoglycerides; wax esters; lipid catabolites; mono-, sesqui-, and straight-chain terpenes; and an aromatic hydrocarbon from the shikimic acid route. Five non-volatile compounds were identified, which have been described in P. roqueforti for the first time, namely, triolein, ergosterol peroxide, 9(11)-DEPH, 4-hydroxybenzaldehyde, and d-mannitol. In summary, an unusual strain of P. roqueforti that did not produce any toxins was found. This study allowed identification of compounds that were already reported in the literature, but it also detected new compounds for this fungus. The cultured strain of P. roqueforti does not contain any component of the volatile chemical components involved in the biogenesis of PR-toxin, which is consistent with the chemical tracers proposed by Demyttenaere et al.,17 Jelen,18 and Calvert et al..19 Moreover, there are no indications the studied strain may produce botryodiplodin, the mycotoxin described by Moreau et al.56 in a P. roqueforti strain that does not produce the PR-toxin. The results of this study support the idea that the metabolic profiles of the mycelia of this P. roqueforti strain can be potentially used as therapeutic agents and natural sources for the production of nutraceuticals and functional foods. Although relatively closely studied, there is no doubt that these halotolerant filamentous fungi still represent an intriguing area of research for the production of new, high-value biomolecules.
SUPPLEMENTARY MATERIAL A detailed descriptive mycelial study together with the 1H and MS spectra of the compounds 15, 26, 27, 28; 13C and TOCSY spectra of the compound 26; HSQC and HMBC spectra of the compounds 27 and 28; and GC-MS fingerprint of the compounds 5 (n = 12, 13, and 18) and 26 can be seen at http://quimicanova.sbq.org.br, in PDF file, with free access. The GC-MS spectra of additional compounds and further spectroscopy data are available on request.
ACKNOWLEDGMENTS The authors would like to thank the CAPES agency (Coordenaçao de Aperfeiçoamento de Pessoal de Nível Superior, Brazil) for the PhD fellowship and the European Commission for a Marie Curie Training Site Fellowship, both granted to Roberto Mioso. We gratefully acknowledge the financial support given to the project SI-697 (ULPAPD-08/01-5) by the Canary government (Agencia Canaria de Investigación, Innovación y Sociedad de la Información, ACIISI) and the ICIC (Instituto Canario de Investigación del Cáncer).The authors also express their thanks to Margaret Hart for revising the English of this manuscript.
REFERENCES 1. Bourdichon, F.; Casaregola, S.; Farrokh C.; Frisvad, J. C.; Gerds, M. L.; Hammes, W. P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; Powell, I. B.; Prajapati, J. B.; Seto, Y.; Schure, E. T.; Boven, A. V.; Vankerckhoven, V.; Zgoda, A.; Tuijtelaars, S.; Hansen, E. B; Int. J. Food Microbiol. 2012, 154, 87. 2. Frisvad, J. C.; Andersen, B.; Thrane, U.; Mycol. Res. 2008, 112, 231. Rasmussen, R. R.; Rasmussen, P. H.; Larsen, T. O.; Bladt, T. T.; Binderup, M. L.; Food Chem. Toxicol. 2011, 49, 31. 3. Kubeczka, K. H.; Archiv. Mikrobid. 1968, 60, 139. Van den Tempel, T.; Nielsen, M. S.; Int. J. Food Microbiol. 2000, 57, 193. 4. Godinho, M; Fox, P. F.; Milchwissenschaft 1981, 36, 205. 5. Vansteelandt, M.; Kerzaon, I.; Elodie Blanchet, E.; FossiTankoua, O.; Robiou du Pont, T.; Joubert, Y.; Monteau, F.; Le Bizec, B.; Frisvad, J. C.; Pouchus, Y. F.; Grovel, O.; Fungal Biol., in press. 6. Engel, G.; Spicher, G.; Getreide, Mehl und Brot 1985, 39, 123.; Engel, G.; Teuber, M.; Milchwissenschaft 1983, 38, 513. Peberdy, J. F.; In Biology of Industrial Microorganisms; Demain, A. L.; Solomon, N. A., eds.; The Benjamin/ Cummings Pub. Co., Inc.: London, UK, 1985.; Sharpell, F. H. Jr.; In Comprehensive Biotechnology; Pergamon Press: New York, 1985. Szoltysek, K; Roczniki PZH 1982, 33, 39. 7. Lee, B. H.; Clothier, M. F.; Dutton, F. E.; Nelson, S. J.; Johnson, S. S.; Thompson, D. P.; Geary, T. G.; Whaley, H. D.; Haber, C. L.; Marshall, V. P.; Gabe, I.; McNally, P. L.; Ciadella, J. I.; Martin, D. G.; Bowman, J. W.; Baker, C. A.; Coscarelli, E. M.; Alexander-Bowman, S. J.; Davis, J. P.; Zinser, E. W.; Wiley, V.; Lipton, M. F.; Mauragis, M. A; Curr. Top.Med. Chem. 2001, 2, 779.; Mrozik, H.; US pat.88-233785 1989. Ruddock, J.; UK pat. GB 90-17922 1992. 8. Kopp-Holtwiesche, B.; Rehm, H. J.; J. Environ. Pathol. Toxicol. Oncol. 1990, 10, 41. 9. Woloszyn, J.; Szoltysek, K.; Ziobrowski, J.; Pol. Przemysl Spozywczy 1988, 42, 297.; Venugopal, V.; Lakshmanan, R.; Doke, S. N.; Bongirwar, D. R.; Food Biotechnol. 2000, 14, 21. 10. Hassanien, F. R.; Ragab, M.; El-Makhzangy, A; Fette, Seifen, Anstrichmittel 1986, 88, 33.; Ragab, M.; Hassanien, F. R.; El-Makhzangy, A.; Fette, Seifen, Anstrichmittel 1986, 88, 72. 11. Lawrence, L.C.; J. Gen. Microbiol. 1967, 46, 65; Salvadori, P.; Salvadori, B. B.; Le Lait 1967, 47, 605; Lawrence, R. C.; Hawke, J. C.; J. Gen. Microbiol. 1968, 51, 289; Fan, T. Y.; Kinsella, J. E.; J. Sci. Food Agric. 1976, 27, 745. 12. Lomascolo, A.; Dubreucq, E.; Perrier, V.; Galzy, P.; Grimaud, J.; J. Dairy Sci. 1994, 77, 2160.; Shimp, J. L.; Kinsella, J. E.; J. Food Sci. 1977, 42, 681. 13. Kaufmann, H. P.; Ahmad, A. K. S.; Radwan, S. S.; Fette, Seifen, Anstrichmittel 1966, 68, 498. 14. Lawrence, L. C.; J. Gen. Microbiol. 1967, 46, 65; Salvadori, P.; Salvadori, B. B.; Le Lait 1967, 47, 605; Lawrence, R. C.; Hawke, J. C.; J. Gen. Microbiol. 1968, 51, 289; Fan, T. Y.; Kinsella, J. E.; J. Sci. Food Agric. 1976, 27, 745. 15. Kanota, K.; Eisei Shikensho Hokoku 1969, 87, 31.; Kanota, K.; Proceedings of the U.S.-Japan First Conf. Toxic Micro-Organisms, Honolulu, Hawaii, 1968; Kolousek, J.; Michalik, S.; Sbornik Ceskoslov. Akad. Zemedel. Ved. 1954, 27, 281. 16. Scott, P. M.; Kennedy, B. P. C.; Harwig, J.; Blanchfield, B. J.; Appl. Environ. Microbiol. 1977, 33, 249. 17. Demyttenaere, J. C. R.; Adams, A.; Van Belleghem, K.; De Kimpe, N.; Konig, W. A.; Tkachev, A. V.; Phytochemistry 2002, 59, 597. 18. Jelen, H. H.; J. Agric. Food Chem. 2002, 50, 6569. 19. Calvert, M. J.; Ashton, P. R.; Allemann, R. K.; J. Am. Chem. Soc. 2002, 124, 11636. 20. Pitt, J. I.; In The Genus Penicillium, Academic Press: London, UK, 1999. 21. Frisvad, J. C.; Smedsgaard, J.; Larsen, T. O.; Samson, R. A.; Stud. Mycol. 2004, 49, 201; Smith, J. E.; Lewis, C. W.; Anderson, J. G.; Solomons, G. L.; In Mycotoxins in human nutrition and health, Eur. Comm. Dir. Gen., Brussels, rep. EUR 16048EN, 1994. 22. Pitt, J. I.; Leistner, L.; In Mycotoxins and Animal Foods; J. E. Smith J. E.; Henderson, R. S., eds.; CRC Press: Boca Raton, USA, 1991. 23. Chang, S. C.; Wei, Y. H.; Wei, D. L.; Chen, Y. Y.; Jong, S. C.; Appl. Environ. Microbiol. 1991, 57, 2581. 24. O'Brien, M.; Nielsen, K. F.; O'Kiely, P.; Forristal, P. D.; Fuller, H. T.; Frisvad, J. C.; J. Agric. Food Chem. 2006, 54, 9268; Edel, V.; Polymerase Chain Reaction in Mycology - an Overview. In Application of PCR in Mycology; Bridge, P. D.; Arora, D. K.; Reddy, C. A.; Elander, R. P., eds.; CAB International: Wallingford, 1998. Bugni, T. S.; Ireland, C. M.; Nat. Prod.Rep. 2004, 21, 143. 25. Masuma, R.; Yamaguchi, Y.;Noumi, M.; Omura, S.; Michio, N.; Mycoscience 2001, 42, 455; Furtado, N. A. J. C.; Said, S.; Ito, I. Y.; Bastos, J.K.; Microbiol. Res. 2002, 157, 207. 26. Seimour, R. L.; Fuller, M. S.;Collection and isolation of water molds(Saprolegniaceae)from water and soil. In Zoosporic fungi in teaching and research; Fuller, M. S.; Jaworowskia, A., eds.; Southestern Publishing: Athens, 1987. 27. Kupchan, S. M.; Briton, R. W.; Ziegler, M. F.; Siegel, C. W.; J. Org. Chem. 1973, 38, 178. 28. Carballeira, N. M.; Miranda, C.; Lozano, C. M.; Nechev, J. T.; Ivanova, A.; Ilieva, M.; Tzvetkova, I.;Stefanov, K.; J. Nat. Prod. 2001, 64, 256; Duncan, W. R. H.; Lough, A. K.; Garton, G. A.; Brooks, P.; Lipids 1974, 9, 669; Rezanka, T.; Nedbalova, L., Elster, J.; Cajthaml, T.; Sigler, K.; Phytochemistry 2009, 70, 655; Rezanka, T.; Sigler, K.; Prog. Lipid Res. 2009, 48, 206. 29. Buckner, J. S.; Kolattukudy, P. E.; Biochemistry 1975, 14, 1774; Seyama, Y.; Oaci, K.; Imamura, T.; Kasama, T.; Otsuka, H.; J. Biochem. 1983, 94, 1231. 30. Cooper, W. J.; Blumer, M.; Deep Sea Res. 1968, 15, 535; Cranwell, P. A.; Chem. Geol. 1973, 11, 307; Cranwell, P. A.; Chem. Geol. 1974, 14, 1; Grimalt, J. O.; Albaigés, J.; Mar. Geol. 1990, 95, 207; Kaneda, T.; Microbiol. Rev. 1991, 55, 288; Leo, R. G.; Parker, P. L.; Science 1966, 152, 649. 31. Anderson, B. A.; Progress in the Chemistry of Fats and other Lipids 1978, 16, 279. 32. Dyal, S.; Narine, S. S.; Food Res. Int. 2005, 38, 445. 33. González, A. G.; Barrera, J. B.; Marante, F. J. T.; Phytochemistry 1983, 22, 1049. 34. Windaus, A.; Brunken, J.; Liebigs Ann. Chem. 1928, 460, 225; Bowen, E. J.; In Advances in Photochemistry; Noyes, W. A.; Hammond, G. S.; Pitts, J. N., eds.; Wiley Interscience: New York, USA, 1963. 35. Sheikh, Y.; Djerassi, C.; Tetrahedron 1974, 30, 4095. 36. Kim, S. W.; Park, S. S.; Min, T. J.; Yu, K. H.; Bull. Korean Chem. Soc. 1999, 20, 819. 37. Duarte, N.; Ferreira, M. J.; Martins, M.; Viveiros, M.; Amaral, L.; Phytotherapy Research 2007, 21, 601. 38. Bok, J. W.; Lermer, L.; Chilton, J.; Klingeman, H. G.; Towers, G. H.; Phytochemistry 1999, 51, 891; Yaoita, Y.; Ebina, K.; Kakuda, R.; Machida, K.; Kikuchi, M.; Nat. Med. 2002, 56, 117; Takei, T.; Yoshida, M.; Ohnishi-Kameyama, M.; Kobori, M.; Biosci. Biotechnol. Biochem. 2005, 69, 212. 39. Marante, F. J. T.; Mioso, R.; Barrera, J. B.; González, J. E. G.; Rodríguez, J. J. S.; Bravo de Laguna, I. H.; Ann. Microbiol. 2012, 62, 1601. 40. Nam, K. S., Jo, Y. S., Kim, Y. H., Hyun, J. W.; Kim, H. W.; Life Sci. 2001, 69, 229. 41. Marinho, A. M. R.; Marinho, P. S. B.; Rodrigues Filho, E.; Quim. Nova 2009, 32, 1710. 42. Fisch, M. H.; Ernst, R.; Flick, B. H.; Arditti, J.; Barton, D. H. R.; Magnus, P. D.; Menzies, I. D.; J. Chem. Soc. Chem. Commun. 1973, 23, 530. 43. Mediavilla, M. J.; PhD Thesis, Universidad de La Laguna, Spain, 1984. 44. Amade, P.; Mallea, M.; Bouaiacha, N; J. Antibiot.1994, 47, 201. 45. Peat, S., Whelau, W. J., Lawdey, H. G.; J. Chem. Soc. 1958, 729. 46. Wiseelink, H. W.; Weusthuis, R. A.; Eggink, G.; Hugenholtz, J.; Grobben, G. J.; Int. Dairy J. 2002, 112, 151. 47. Lewis, D.; Smith, D.; New Phytol. 1967, 66, 143. 48. Stoop, J. M. H.; Mooibroek, H.; Appl. Environ. Microbiol. 1998, 64, 4689;Ruijter, G. J.; Bax, M.; Patel, H.; Flitter, S. J.; van de Vondervoort, P. J.; de Vries, R. P.; vankuyk, P. A.; Visser, J.; Eukaryotic Cell 2003, 2, 690. 49. Soetaert, W.; Vanhooren, P. T.; Vandamme, E. J.; Methods in Biotechnology 1999, 10, 261; Makkee, M.; Kieboom, A. P. G.; Van Bekkum, H.; Starch 1985, 37, 136. 50. Itoh, Y.; Tanaka, A.; Araya, H.; Ogasawara. K.; Inabi, H.; Sakamoto, Y.; et al.; EU pat. 486 024 1992. 51. Oliveira, P. S. M.; Ferreira, V. F.; de Souza, M. V. N.; Quim. Nova 2009, 32, 441. 52. Von Weymarn, N.; Hujanen, M.; Leisola, M.; Process Biochemistry 2002, 37, 1207. 53. Archer, D. B.; Current Opinion in Biotechnology 2000, 11, 478. 54. Abbas, A.; Dobson, A. D. W.; In Encyclopedia of Dairy Sciences, 2nd ed., 2011; Fujimoto, H.; In Encyclopedia of Dairy Sciences, 2nd ed., 2011. 55. Frisvad, J. C.; Filtenborg, O.; Mycologia 1989, 81, 836; Paterson, R. R. M.; Venâncio, A.; Lima, N.; Revista Iberoamericana de Micología 2006, 23, 155. 56. Moreau, S.; Lablanche-Combier, A.; Biguet, J.; Foulon, C.; Delfosse, M.; J. Org. Chem. 1982, 47, 2358. |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access






