Artigo
| Binary co-culture selection from marine-derived microorganisms for differential production of specialized metabolites |
|
Paola A. Martínez-Buitrago; Freddy A. Ramos; Leonardo Castellanos*
Universidad Nacional de Colombia, Sede Bogotá, Departamento de Química, Carrera 30 # 45-03, Bogotá D.C. 111321, Colombia Recebido em 18/01/2019 *e-mail: lcastellanosh@unal.edu.co Nowadays the discovery of new compounds from microorganisms has substantially decreased. Genomic studies have shown that certain genes are not expressed under standard laboratory conditions. In order to overcome this problem, several methods including microorganism co-culture have been developed. The aim of this work was to select binary co-cultures of marine-derived microorganisms to pursue further chemical studies. From 15 microorganisms, 151 interactions assays in solid media were performed using two different methodologies: distance assays and contact assays that allowed to distinguish between interactions due to diffusible compounds or due to cell-cell contact. For distance assays, the metabolic production changes were evaluated using 3 zones: two corresponding to each microorganism colony, and a third zone corresponding to the interaction zone. This division zone strategy allowed to assess the diffusion compounds gradient, and to suggest the producer microorganism of the induced compounds. The metabolic production of the co-cultures was evaluated using HPLC-DAD and NMR. Seven co-cultures were identified to be able to induce changes in the metabolic production. Remarkably, the co-culture between the fungus Purpureocillium sp. PNM-67 and the bacteria Rhodococcus sp. RKHC-26 or Gordonia sp. PNM-25 induce production of a red dye in the fungus. These are the first successful examples of interaction between a fungus and a mycolic acid-containing bacteria. INTRODUCTION Over the years, compounds isolated from microorganisms have played an important role in the treatment of human diseases and have revolutionized the pharmaceutical industry. However, redundant isolation of microbial compounds is increasingly common.1 Recent research on microorganisms' genome has shown that there are silent and cryptic biosynthetic pathways, which are not expressed in standard laboratory growth conditions. To promote the activation of these routes, different methods have been developed, including microorganism co-culture.2 This approach takes into account that microorganisms normally grow in complex environments, in this way inter- or intraspecies interactions are prevalent in nature, and the growth of a microorganism in the presence of others, can alter or stimulate their specialized metabolism.3 In fact, some studies have shown that when microorganisms interact with others, they produce specialized metabolites that can have positive (e.g. communication with other microorganisms) or negative (e.g. antibiotics) effects on others.4,5 Both scenarios are equally interesting on the discovery of novel compounds, and on the understanding of the role of specialized metabolites in nature. Microorganism co-culture consists in the cultivation of two or more microorganisms in the same confined environment.6 One of the first successful examples of co-cultivation was reported by Cueto and collaborators in 2001.7 In this study, a co-culture between the fungus Pestalotia sp. and the bacteria Thalassospira sp. was evaluated for its metabolic production, and led to the isolation and structural elucidation of pestalone, a benzophenone that showed antibacterial activity, and whose production was only associated with co-culture. Since then, studies have spread to bacteria-bacteria,8-10 fungus-fungus11-13 and bacteria-fungus14-16 interactions. As a result of co-culture, increased metabolite yields have been reported, as well as the production of analogues of known metabolites and the induction of new bioactive metabolites.17 One of the challenges of using this co-culture approach is the microorganisms selection for co-culture establishment. In most of the reports, the selected microorganisms come from a screening of multiple pairs of microorganisms, or the authors do not mention the reason that support their choice. A revision of specialized literature allowed us to identify the following criteria to select the microorganisms: (1) the use of mycolic acid containing bacteria,18 (2) the establishment of interactions already observed in nature (ecological criteria),19 (3) the confrontation with pathogens,20 (4) the confrontation between bacteria of the genus Streptomyces21 and (5) the confrontation between bacteria of the genus Streptomyces with a bacteria from the phylum Proteobacteria.22 Other challenges in co-culture studies include the selection of factors such as culture media, the degree of contact between microorganisms, population density and fermentation time.23 The degree of contact between the microorganisms showed to be important because the effects can be the result of two types of interaction: interaction due to diffusible compounds (related to nutrient supply or production of an antibiotic or signaling molecule) and interaction due to cell-cell contact.3 Taking this into account, in co-cultures, microorganisms may be perfectly mixed or partially separated. About the culture growth medium, solid or liquid broth can be used. The selection of the culture medium must take into account the two microorganisms involved.23 The solid medium has been used in co-cultures because, unlike the liquid broth, it enables to visualize the morphology and interaction patterns between microorganisms, that allow to observe phenotypic changes including developmental defects, growth inhibition, lysis, motility, pigment production, among others.24 It also facilitates the spatial location of metabolites,25 which allows the identification of the organism responsible for the production of compounds induced by interaction. Although co-cultivation is a promising strategy that has produced successful results, these have generally been a consequence of serendipity.26 For this reason, this strategy still presents disadvantages such as the lack of reproducibility and predictability. To address these drawbacks, this strategy still faces the challenge of developing standardized protocols, as well as establishing methodologies to evaluate a large number of interactions simultaneously.26 This could result in more specific criteria for the selection of microorganisms, or in the characterization of promoting factors produced by stimulating strains that could guide to the development of effective co-culture conditions. In this study, a screening of binary interactions was performed in solid media using different methodologies in order to prioritize microorganisms' co-cultures for the evaluation of their metabolic production.
EXPERIMENTAL SECTION General Bacteriological agar (Scharlau), tryptose (Scharlau), sodium chloride (Merck), yeast extract (Oxoid), malt extract (Scharlau), glucose (Merck), potato dextrose agar (Scharlau), iron sulphate heptahydrate (Chemie Loba), manganese chloride tetrahydrate (Chemie Loba), zinc sulphate heptahydrate (Chemie Loba) and commercial oatmeal were used for culture media. Ethyl acetate (Panreac AppliChem) was used for extractions. For chromatographic analysis, acetonitrile HPLC grade (Honeywell) and deionized water were used. For nuclear magnetic resonance analysis CDCl3 with deuteration grade 99.8 % (Merck) and methanol-d4 with deuteration grade 99.8% (Merck) were used. Microorganism selection Based on criteria reported in the literature, 15 microorganisms (14 bacteria and 1 fungus) were selected from our in-home microorganism collection. This collection of microorganisms was built by the recovery of bacteria and fungi from samples of marine environments including sediments, invertebrates and algae collected at the reef of the island of Providencia and Santa Catalina.27 Primary selection of couples to be evaluated In order to reduce the number of couples to be evaluated in bacteria-bacteria distance interactions, a first assay was carried out to prioritize those couples that showed phenotypic changes. The methodology developed by Seyedsayamdost et al. was adapted.28 Each bacteria was grown in 10 mL of Luria-Bertani medium (composition per liter: tryptose 10 g, NaCl 10 g, yeast extract 5 g) until reaching a OD600nm between 0.6 and 0.8. From each of these cultures 1µL aliquots were taken and sown in 150*15 mm Petri dishes containing solid Luria-Bertani medium (composition per liter: tryptose 10 g, NaCl 10 g, yeast extract 5 g, agar 14 g). In each dish the interaction of 36 pairs was evaluated arranging them in 3 columns separated by 1 cm of distance. In each couple the bacteria were sown 5 mm away. At the left and right end of the Petri dish a column of each single bacteria was sown as control. The seeding arrangement of the bacteria is shown in Figure 1S of the supplementary material. Each pair was evaluated by triplicate. Petri dishes were incubated at 28 ºC for 5 days. After this time, the phenotypic changes induced by co-culture (growth inhibition, pigmentation, sporulation, macroscopic morphology) were evaluated by visual comparison with the respective controls. Binary co-cultures Two different methods were used: a distance assay and a contact assay. The methodology was divided into two types of interactions: bacteria-bacteria and bacteria-fungus. All tests were performed on 90 mm*15 mm Petri dishes. For bacteria the used inoculum was 105 CFU per dish, and in the case of the fungus 105 conidia per dish were used. Bacteria-bacteria interactions Luria-Bertani (composition per liter: tryptose 10 g, NaCl 10 g, yeast extract 5 g, and agar 14 g) was used as culture media. In the distance assay the microorganisms were sown at a distance of 5 mm from each other. For the contact assay, aliquots of each one of the microorganisms were mixed, and this mixture was inoculated on the surface of the agar. The seeding arrangement of the bacteria is shown in Figure 2S of the supplementary material. Three replicates were made of each co-culture. Monocultures of each of the bacteria were performed also by triplicate under the same conditions. In both cases, the dishes were incubated at 30 ºC for 5 days. Bacteria-fungus interactions Cultures were made in solid medium using 4 different culture media: a) LB supplemented with glucose (composition per liter: tryptose 10 g, NaCl 10 g, yeast extract 5 g, glucose 20 g, and agar 14 g), b) PDA (composition per liter: potato peptone 4 g, glucose 20 g, and agar 15 g), c) ISP2 (composition per liter: malt extract 10 g, yeast extract 4 g, glucose 4 g, and agar 20 g), and d) ISP3 (composition per liter: Oatmeal 20 g, agar 18 g, and trace solution 1 mL). For the distance assay, the bacteria were inoculated in the centre of the dish in a strip 1 cm wide, while the fungus was inoculated at opposite ends of the dish. For the contact assay, aliquots of each one of the microorganisms were mixed, and this mixture was then inoculated. The seeding arrangement of microorganisms is shown in Figure 3S of the supplementary material. three replicates of each co-culture were made. Monocultures of each of the bacteria and fungus were made by triplicate under the same conditions as the co-cultures. In both cases, the dishes were incubated at 30 ºC for 10 days. In all established binary co-cultures, phenotypic changes induced by co-culture were visually assessed looking for changes in growth, pigmentation, sporulation, and macroscopic morphology. Extraction In contact assays the agar was cut into 1 cm*1 cm pieces using a sterile blade. In the distance assays, three different zones were previously defined, which can be seen in figure 4S of the supplementary material, and then each zone was cut into pieces separately. Each sample was freeze-dried using a LABCONCO lyophilizer (Freezone 4.5). The dry material was extracted twice with ethyl acetate, at a ratio of 70 mL of solvent per dish. The extractions were performed using a Cole-Parmer ultrasound bath at room temperature for 20 minutes. The samples were filtered and dried at reduced pressure. Metabolic profiling Organic extracts were analyzed by liquid chromatography using a UHPLC Thermo Dionex Ultimate 3000 (Germering) device with a DAD detector. The extracts were previously dissolved in methanol to have a final concentration of 2 mg mL-1, and an injection of 20 µL was made. A Kinetex 2.6 µm C8 (100x4.6 mm) column was used. Solvents used in elution were: water (A) and acetonitrile (B). The analysis program for all the extracts was: a gradient that started with 10% of phase B, and increased linearly to 100% in 23 minutes, keeping this ratio for 2 more minutes. The flow was set to 0.5 mL min-1. The spectra of 1H NMR and COSY for organic extracts were obtained in a Bruker Avance 400, using as solvents CDCl3 and methanol-d4. Differences in the metabolic production of co-cultures, in comparison to monocultures and culture medium, were determined by comparison of HPLC-DAD chromatograms and 1H NMR spectra.
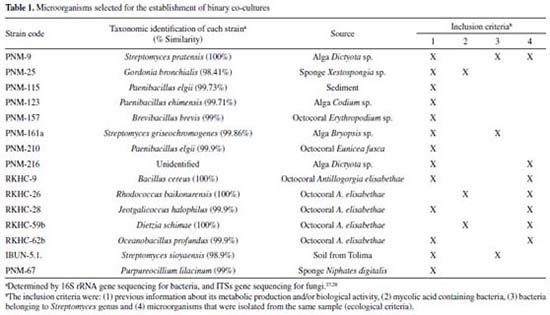
RESULTS AND DISCUSSION Co-culture as a tool of induction of bioactive metabolites is a relatively recent method, without not fully defined criteria for its establishment.26 Some selection criteria for establish the most promising interactions have been described in the literature, which are particularly true for bacteria-bacteria interactions, while for bacterial-fungus interactions this is limited to ecological criteria. In this study, 14 bacteria and 1 fungus were selected from our collection of microorganisms that comprises mostly marine recovered isolates, as well as some soil recovered isolates, which are summarized in Table 1 where the inclusion criteria is also described.
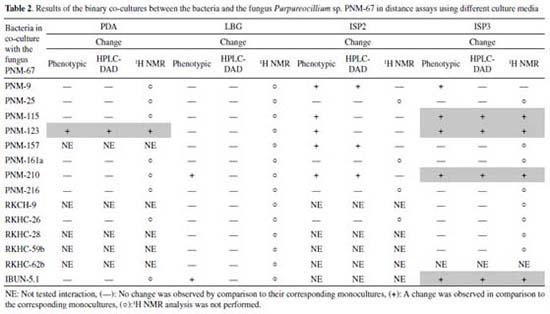
Bacteria-bacteria co-cultures Distance assays Co-cultures assays enable to evaluate effects mediated by diffusible compounds. A first assay that would prioritize couples that would later be evaluated in individual Petri dish assays was conducted. For this purpose, the methodology proposed by Seyedsayamdost and collaborators29 was adapted to evaluate the phenotypic changes associated to interactions, taking into account that these could be indicators of changes in metabolic production. Phenotypic changes were observed in 27 out of 91 tested couples. Growth inhibition was observed in 18 out of the 27 cases and pigmentation changes and/or sporulation in 9 out of the 27 cases (detailed results are found in Figure 5S). These 27 interactions that showed phenotypic changes were cultured in 9cm Petri dishes by triplicate, in order to obtain organic extracts that allowed evaluating the metabolic production. Each extract was analyzed by HPLC-DAD (in the range of 200-600 nm); however, no changes were observed in the metabolic production of 27 co-cultures in comparison with those of the corresponding monocultures. The 18 co-cultures that showed bacterial growth inhibition were analyzed by NMR. This technique did not showed changes in co-cultures spectra in comparison with those of the monocultures spectra. Thus, this methodology did not allow to select any co-cultures to evaluate their metabolite production. It is worth to mention that co-cultures could be producing compounds in response to the presence of the other microorganism, but the analytical techniques here employed did not allow to detect such production changes. More sensitive analytical techniques such as HPLC-MS, could help to detect metabolic production changes in these distance assays.12,30 The above since this technique is several orders of magnitude more sensitive than NMR.31 Also, it is important to consider that while HPLC-MS is more sensitive, their isolation could be difficult because they can be found in small quantities, whereas compounds detected by NMR could be isolated by traditional methods. Contact assays For the selection of couples in this type of assay, there is no applicable methodology such as the phenotypic change assay performed in distance assays. For this reason, some bacteria were not considered in this study, the exclusion criteria are explained below. Co-culture couples containing the bacteria Gordonia sp. PNM-25 and Dietzia sp. RKHC-59b were discarded because their growth rate in the selected culture medium was significantly slower in comparison with other strains. Co-culture couples including Brevibacillus sp. PNM-157, Paenibacillus sp. PNM-210, Paenibacillus sp, PNM-115 and Paenibacillus sp. PNM-123 were also discarded since in previous studies they showed potent antibiotic properties. In order to reduce the number of interactions to be evaluated, 3 criteria derived from literature were used: (1) co-cultures involving 2 strains of the genus Streptomyces; (2) co-cultures involving strains of the genus Streptomyces and mycolic-acid containing bacteria; and (3) co-cultures involving 2 strains obtained from the same environmental sample. In this way, 13 selected pairs were grown in 9cm petri dishes by triplicate. Co-cultures results are summarized in Table 1S of the supplementary material. The 1H NMR analyses of these 13 co-cultures allowed to identify Streptomyces sp. PNM-161a and Rhodococcus sp. RKHC-26 co-culture as producer of compounds absent in both, the broth blank and the monocultures. The signals for induced compounds are found at δH 1.5-1.7, 2.35-2.50, and 2.95 (bt, J=7.5Hz) for aliphatic protons, and at δH 7.15 (m) for aromatic protons (Figure 6S). Bacteria-fungus co-cultures Most of successful co-culture examples involve at least one fungus.32 Thus, pairings between the fungal isolate Purpureocillium sp. PNM-67 and 14 different bacteria were performed. As initial criterion for matching, the capability of growing in four different culture media was used (Table 2). All co-cultures were initially evaluated in a distance assay, and those couples that had a phenotypic change when the two microorganisms came into contact, were also evaluated in a contact assay. The results of both approaches are summarized below.
Distance assays Distance assays were performed on 4 different culture media (PDA, LBG, ISP2, ISP3). A total of 45 assays were conducted and phenotypic changes were observed in 13 cases. The predominant phenotypic change was inhibition of fungal growth in response to its interaction with several bacteria. In order to obtain the organic extracts from these assays, three zones at Petri dish were defined, the first one where the fungus was inoculated; the second one where the bacteria was inoculated; and a third zone, which will be called “interaction zone” because it was spatially between the two inoculums. This division allowed to suggest which was the producer of the compounds due to their diffusion gradient. The obtained results are summarized in Table 2. Phenotypic changes were observed in 13 out of 45 assays; in all cases inhibition of fungal growth was observed. Extracts from the interaction zones of these 13 co-cultures were analyzed by HPLC-DAD, and changes were observed in 8 cases (1 case in PDA, 3 in ISP2 and 4 in ISP3). It is important to observe that the bacteria involved in these co-cultures belong to the genera Streptomyces sp. (PNM-9 and IBUN-5.1.), Paenibacillus sp. (PNM-115, PNM-123 and PNM-210) and Brevibacillus sp. (PNM-157). These 8 co-cultures were also analyzed by 1H NMR, and changes were found in 5 cases which are described below. In the first co-culture, between the fungus Purpureocillium sp. PNM-67 and the bacterium Paenibacillus sp. PNM-123 in PDA; changes in phenotype, in HPLC-DAD and 1H NMR were observed. HPLC analysis for the interaction zone, showed two peaks (maximum absorbance at 320 nm) that are not present in the monocultures. These peaks are also present in the bacteria zone in the co-culture dish, so the diffusion gradient of these compounds indicates that they are produced by the bacteria in response to its interaction with the fungus. This co-culture was also analyzed by 1H NMR, where signals exclusive to co-culture extracts in bacterial and interaction zones were found between 7.6 and 7.8 ppm. Changes induced by co-culture are showed in Figure 1. Both analytical techniques suggest that compounds induced by co-culture have aromatic features.
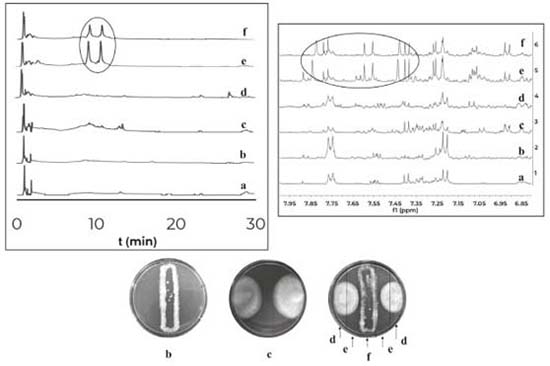 Figure 1. Comparison of HPLC-DAD chromatograms (320nm) at the left and comparison of 1H NMR spectra at the right for the organic extracts of: (a) culture medium PDA, (b) monoculture of Purpureocillium sp. PNM-67, (c) monoculture of Paenibacillus sp. PNM-123 and the three zones of the co-culture dish (d) fungus zone, (e) interaction zone and (f) bacteria zone
The co-cultures of the fungus PNM-67 with the bacteria Paenibacillus sp. PNM-115, Paenibacillus sp. PNM-123, Paenibacillus sp. PNM-210 and Streptomyces sp. IBUN-5.1, in medium ISP3 showed by HPLC one same peak (both in retention time and UV spectrum) in all cases that can be associated to co-culture. This peak was obtained from interaction zone of the co-culture extracts and was absent in the corresponding monocultures. The 1H NMR analyses for these extracts showed the presence of the same signals but in different proportions (Figure 2).
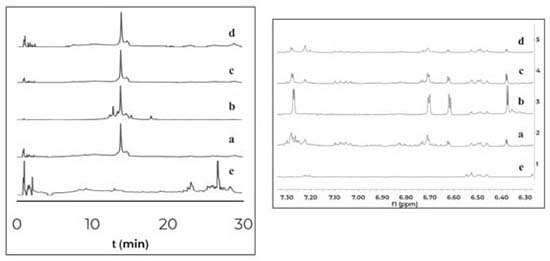 Figure 2. Comparison of HPLC-DAD chromatograms (320nm) at the left and 1H NMR spectra (expansion 6-8ppm) at the right for organic extracts of the interaction zones of the co-cultures between Purpureocillium sp. PNM-67 and: (a) Paenibacillus sp. PNM-123, (b) Paenibacillus sp. PNM-115, (c) Streptomyces sp. IBUN-5.1 and (d) Paenibacillus sp. PNM-210. (e) Monoculture of the fungus Purpureocillium sp. PNM-67
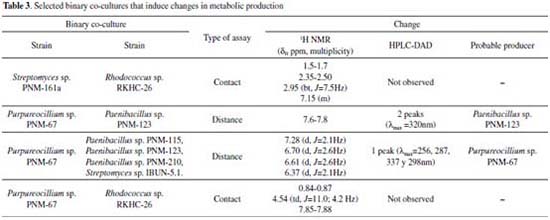
The results indicate that the induced compounds were produced by the fungus because the signals (HPLC-DAD and 1H NMR) were found in both, the interaction zone and the fungus zone of the 4 co-cultures. In addition, the possible role of these compounds is the defense of the fungus against the bacteria, taking into account that there is inhibition of fungal growth in the 4 co-cultures. Using the COSY experiment, it was found that the signals at 7.28 and 6.37 ppm coupled among them, indicating that they are part of the same compound. Further studies will enable to establish if the signals in NMR and the peak in HPLC-DAD correspond to the same compound(s). Contact assays In distance assays, two co-cultures stood out due to their reddish pigmentation that happened when the fungus came into contact with the bacteria Gordonia sp. PNM-25 as well as with Rhodococcus sp. RKHC-26. It is noteworthy that these two bacteria were initially included in the set of microorganisms because they are mycolic acid-containing bacteria. In the case of bacteria-bacteria co-cultures, bacteria containing mycolic acid have been shown to be able to induce the production of specialized metabolites in bacteria from the Streptomyces genus.33 Recent studies have shown that the effect of mycolic acid containing bacteria also extends to other Actinobacteria;34 however, there are no reports showing that they also have such effect on fungus35. In this way, these two co-cultures could be of great interest, as they constitute the first examples of the effect of mycolic acid containing bacteria on fungi. The fact that these two co-cultures show changes only when performed by a contact assay methodology support the reports that show that mycolic acid containing bacteria exert their effect only when they are alive and have cell to cell contact with the other organism.35 In an effort to increase the production of this red pigment, these two co-cultures were evaluated in a contact assay. In the case of the co-culture between PNM-67 and RKHC-26, the red pigment could be observed in the whole dish.On the other hand Gordonia sp. PNM-25 did not survive to the assay conditions. Changes to the methodology of the contact assay (like inoculation timing) could lead to the survival of Gordonia sp. PNM-25. There are many reports of microbial pigments functions, ranging from their protective activity against ultraviolet radiation, antimicrobial oxidants or compounds produced by other microorganisms, until its antimicrobial activity against other microorganisms.36 In addition, the production of this red pigment can be used as an indicator of change in another metabolites production.28,33,37 HPLC-DAD analyses showed no single peak for co-culture while 1H NMR analyses showed signals in δH 0.84-0.87, 4.54 (td, J=11.0; 4.2 Hz), and 7.85-7.88 exclusive for co-culture, as is showed in Figure 3.
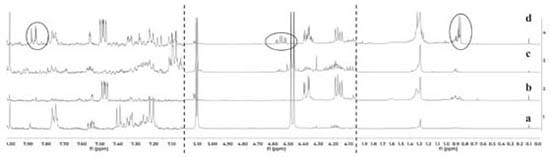 Figure 3. Comparison of 1H NMR spectra for organic extracts of a) culture medium LB, (b) monoculture of Purpureocillium PNM-67, (c) monoculture of Rhodococcus sp. RKHC-26 and (d) co-culture of them
The most relevant information of the selected co-cultures during this study is summarized in Table 3.
CONCLUSIONS From a set of 15 microorganisms (14 bacteria and 1 fungus), co-cultures between bacteria-bacteria and bacteria-fungus were evaluated through 151 assays, using distance interactions and contact interactions in solid media. The main advantage of distance assays over contact assays is that the former facilitates the survival of the two strains. Seven co-cultures induce changes in the metabolic production, five of them due diffusible compounds, and two of them due to cell-to-cell contact. The use of HPLC-DAD and NMR techniques, as a strategy for monitoring metabolic production, can be used to detect and monitor larger changes in metabolic profile, with the advantage that this technique gives information about the isolable compounds. However, the use of LC-MS/MS for co-culture monitoring could be a valuable technique in order to follow small metabolic changes related to interactions. The co-culture between the fungus Purpureocillium sp. PNM-67 and the bacteria Rhodococcus sp. RKHC-26 or Gordonia sp. PNM-25 induce red pigmentation in the fungus Purpureocillium sp. PNM-67, that could be associated with a change in the metabolic production. These are the first successful examples of interactions between a fungal isolate and a mycolic acid-containing bacterium. This kind of interaction could be considered as criteria for selecting couples for co-culture studies.
SUPPLEMENTARY MATERIAL The figures related to the experimental part and some details about the results obtained are in the supplementary material that is available in http://quimicanova.sbq.org.br in the form of an open PDF file.
ACKNOWLEDGMENTS P. Martínez thanks to Colciencias for the program “Jóvenes Investigadores Convocatoria 761 de 2016”. This work was supported in part by research grants from Colciencias and Universidad Nacional de Colombia-DIB. We would like to thank to ANLA and Ministerio de Ambiente y Desarrollo Sostenible for the permission to collect samples and perform this research (Permiso No. 4 de 10/02/2010, Anexo 2, Contrato de Acceso a Recurso Genético No. 108 Otrosi No. 6 al contrato marco de acceso a recurso genético y sus productos derivados No. 121 del 22 de enero del 2016).
REFERENCES 1. Katz, L.; Baltz, R. H.; J. Ind. Microbiol. Biotechnol. 2016, 43, 155. 2. Ochi, K.; J. Antibiot. (Tokyo) 2017, 70, 25. 3. Okada, B. K.; Seyedsayamdost, M. R.; FEMS Microbiol. Rev. 2017, 41, 19. 4. Abrudan, M. I.; Smakman, F.; Grimbergen, A. J.; Westhoff, S.; Miller, E. L.; van Wezel, G. P.; Rozen, D. E.; Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 11054. 5. de Weert, S.; Kuiper, I.; Lagendijk, E. L.; Lamers, G. E. M.; Lugtenberg, B. J. J.; Mol. Plant-Microbe Interact. 2004, 17, 1185. 6. Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L.; Biotechnol. Adv. 2014, 32, 1180. 7. Cueto, M.; Jensen, P. R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J.; J. Nat. Prod. 2001, 64, 1444. 8. Dashti, Y.; Grkovic, T.; Abdelmohsen, U.; Hentschel, U.; Quinn, R.; Mar. Drugs 2014, 12, 3046. 9. Patin, N. V.; Floros, D. J.; Hughes, C. C.; Dorrestein, P. C.; Jensen, P. R.; Microbiology 2018, 164, 946. 10. Adnani, N.; Chevrette, M. G.; Adibhatla, S. N.; Zhang, F.; Yu, Q.; Braun, D. R.; Nelson, J.; Simpkins, S. W.; McDonald, B. R.; Myers, C. L.; Piotrowski, J. S.; Thompson, C. J.; Currie, C. R.; Li, L.; Rajski, S. R.; Bugni, T. S.; ACS Chem. Biol. 2017, 12, 3093. 11. Bertrand, S.; Schumpp, O.; Bohni, N.; Monod, M.; Gindro, K.; Wolfender, J.-L.; J. Nat. Prod. 2013, 76, 1157. 12. Bohni, N.; Hofstetter, V.; Gindro, K.; Buyck, B.; Schumpp, O.; Bertrand, S.; Monod, M.; Wolfender, J.-L.; Molecules 2016, 21, 370. 13. Serrano, R.; González-Menéndez, V.; Rodríguez, L.; Martín, J.; Tormo, J. R.; Genilloud, O.; Front. Microbiol. 2017, 8, 649. 14. Yu, L.; Ding, W.; Wang, Q.; Ma, Z.; Xu, X.; Zhao, X.; Chen, Z.; Tetrahedron 2017, 73, 907. 15. Wakefield, J.; Hassan, H. M.; Jaspars, M.; Ebel, R.; Rateb, M. E.; Front. Microbiol. 2017, 8, 1284. 16. Wu, C.; Zacchetti, B.; Ram, A. F. J.; van Wezel, G. P.; Claessen, D.; Hae Choi, Y.; Sci. Rep. 2015, 5, 10868. 17. Pettit, R. K.; Appl. Microbiol. Biotechnol. 2009, 83, 19. 18. Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T.; Appl. Environ. Microbiol. 2011, 77, 400. 19. Rateb, M. E.; Hallyburton, I.; Houssen, W. E.; Bull, A. T.; Goodfellow, M.; Santhanam, R.; Jaspars, M.; Ebel, R.; RSC Adv. 2013, 3, 14444. 20. Burgess, J. G.; Jordan, E. M.; Bregu, M.; Mearns-Spragg, A.; Boyd, K. G.; J. Biotechnol. 1999, 70, 27. 21. Ueda, K.; Kawai, S.; Ogawa, H.; Kiyama, A.; Kubota, T.; Kawanobe, H.; Beppu, T.; J. Antibiot. (Tokyo) 2000, 53, 979. 22. Carlson, S.; Tanouye, U.; Omarsdottir, S.; Murphy, B. T.; J. Nat. Prod. 2015, 78, 381. 23. Goers, L.; Freemont, P.; Polizzi, K. M.; J. R. Soc. Interface 2014, 11, 20140065. 24. Stubbendieck, R. M.; Vargas-Bautista, C.; Straight, P. D.; Front. Microbiol. 2016, 7, 1234. 25. Yang, Y.-L.; Xu, Y.; Straight, P.; Dorrestein, P. C.; Nat. Chem. Biol. 2009, 5, 885. 26. Nai, C.; Meyer, V.; Trends Microbiol. 2018, 26, 538. 27. Betancur, L. A.; Naranjo-Gaybor, S. J.; Vinchira-Villarraga, D. M.; Moreno-Sarmiento, N. C.; Maldonado, L. A.; Suarez-Moreno, Z. R.; Acosta-González, A.; Padilla-Gonzalez, G. F.; Puyana, M.; Castellanos, L.; Ramos, F. A.; PLoS One 2017, 12, e0170148. 28. Correa, H.; Haltli, B.; Duque, C.; Kerr, R.; Microb. Ecol. 2013, 66, 972. 29. Seyedsayamdost, M. R.; Traxler, M. F.; Clardy, J.; Kolter, R.; Methods Enzymol. 2012, 517, 89. 30. Caraballo-Rodríguez, A. M.; Dorrestein, P. C.; Pupo, M. T.; Sci. Rep. 2017, 7, 5373. 31. Wolfender, J.-L.; Planta Med. 2009, 75, 719. 32. Adnani, N.; Rajski, S. R.; Bugni, T. S.; Nat. Prod. Rep. 2017, 34, 784. 33. Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T.; Appl. Environ. Microbiol. 2011, 77, 400. 34. Hoshino, S.; Okada, M.; Awakawa, T.; Asamizu, S.; Onaka, H.; Abe, I.; Org. Lett. 2017, 19, 4992. 35. Hoshino, S.; Onaka, H.; Abe, I.; J. Ind. Microbiol. Biotechnol. 2018, 1. 36. Liu, G. Y.; Nizet, V.; Trends Microbiol. 2009, 17, 406. 37. Yang, Y.-L.; Xu, Y.; Kersten, R. D.; Liu, W.-T.; Meehan, M. J.; Moore, B. S.; Bandeira, N.; Dorrestein, P. C.; Angew. Chem., Int. Ed. 2011, 50, 5839. |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access