Artigo
| In silico prediction of inhibitory potential of a punicalagin β-anomer against SARS CoV-2 main protease (Mpro) |
|
Norberto MonteiroI,*; Vitória MonteiroII; Lorena LimaII; Anna KarollineII; Richele MachadoII
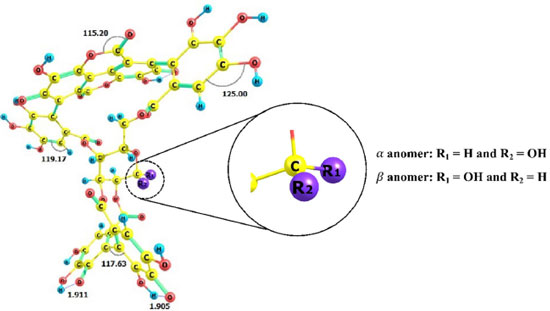
I. Departamento de Química Analítica e Físico-Química, Universidade Federal do Ceará, Campus do Pici, 60440-900 Fortaleza - CE, Brasil Recebido em: 22/02/2022 *e-mail: norbertokv@ufc.br The pandemic caused by the new coronavirus has resulted in a global health emergency and has prompted an urgent need for new treatment strategies. No target-specific drugs are currently available for SARS-CoV-2, but new drug candidates targeting the viral replication cycle are being explored. A prime target of drug-discovery efforts is the SARS-CoV-2 main protease (Mpro). In this work, we identified a potential inhibitor for SARS-CoV-2 main protease using in silico methodologies. Molecular docking and molecular dynamics studies were carried out to ascertain the inhibitory action of α and β anomers of Punicalagin from fruit peel of Punica granatum against the Mpro protease. The molecular dynamics results revealed that the β-anomeric configuration of punicalagin allowed access to more hydrogen bonds and hydrophobic interaction leading to higher selectivity and specificity of β-anomer than α-anomer. Therefore, the β-anomer of Punicalagin could act as potential inhibitor against the main protease of SARS-CoV-2 and may act as a potential drug candidate. INTRODUCTION The outbreak caused by a new species of coronavirus led the World Health Organization (WHO) in early 2020 to declare the disease caused by this virus as a Public Health Emergency of International Importance and soon after it was characterized as a pandemic.1 In December 2019, in the city of Wuhan, China, an outbreak occurred, in which several individuals went to local hospitals with severe pneumonia of unknown etiology. After investigations to find out about the cause of this outbreak, it was found that most of the initial cases had contact with the same wholesale seafood market.2 The etiologic agent discovered was a new virus of the family Coronaviridae and genus β-coronavirus and was identified as severe acute respiratory syndrome of coronavirus 2 (SARS-CoV-2), which causes coronavirus 2019 or COVID-19, as it became known. In addition, it was shown that, unlike other coronaviruses that infect humans, SARS-CoV-2 has a high human-to-human transmission rate, which was a determining factor for its dissemination.3,4 The virus can infect individuals of all ages, however it mainly affects men over 60 years.5 According to He, Deng and Li,6 the lethality rate is higher in patients with coexisting medical conditions, such as diabetes, hypertension and cardiovascular diseases. Symptoms vary from fever, cough, sore throat, changes in smell and taste to acute respiratory distress syndrome (ARDS) and dyspnoea in the most severe cases.6-8 The SARS-CoV-2 virus consists of four structures, the spike glycoprotein or S glycoprotein, a membrane protein, an envelope protein and a nucleocapsid protein, which is present within the viral envelope.9 SARS-CoV-2 is a ribonucleic acid (RNA) virus whose genetic material is a single positive RNA molecule. The virus enters the human host, through the binding of the spike glycoprotein, in the S1 subunit, with its receptor, the angiotensin-converting enzyme 2 (ACE2) that is present on the cell surface.10 After binding, Furin enzyme will pre-cleave the S glycoprotein in the S1 and S2 subunits and then the transmembrane serine protease 2 (TMPRSS2) enzyme will cleave the S glycoprotein in the S2 subunit. Subsequently, the S2 subunit will assist in the fusion of the viral membrane with the host cell membrane and thus, with the entry of the virus into the host cell.4,11 Therefore, the mechanism described above suggests that any organ that overexpresses ACE2 is susceptible to infection to SARS-CoV-2. Covid-19 patients with diarrhea, kidney and testicle damage, intestinal inflammation and pancreatitis were reported to be related with overexpression of ACE2 in the organs related to these diseases.12-15 Though ACE2 is mainly expressed in human type II alveolar cells (AT2), the proportion of type II cells compared to type I is very low, about 1.4% of all AT2 cells.16 Due to this low percentage, it is possible that there is another mechanism that involves SARS CoV-2 infection that explain its high infectivity. In this way, CD147/Basigin,17 neuropilin receptors,18,19 and heparin sulfate20 have already been reported to facilitate SARS-CoV-2 infection. There is a great effort from researchers to find new drugs to combat the SARS-CoV-2 virus. Antiviral for Ebola, remdesivir, has been approved by US Food and Drugs Administration (FDA) and was shown to be efficient in inhibit SARS-CoV-2 virus.21 The remdesivir acts as a nucleoside analog and inhibits the RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 virus. Remdesivir is incorporated by the RdRp and allows for addition of three more nucleotides before RNA synthesis stalls.22 Currently, researchers are looking for several ways to alleviate the severity of the disease and reduce its spread, aiming at a better quality of life after the pandemic.23 Therefore, the search for new compounds that act by inhibiting the SARS-CoV-2 virus infection by different mechanisms is still indispensable. The SARS-CoV-2 main protease is an enzyme called Mpro or 3C-like protease (3CLpro), that plays an important role in the replication cycle, thus becoming a potential target for the control of viral infection.24 Its action occurs after the virus enters the host cells, where the viral RNA genome will translate inactive proteins, called pp1a and pp1ab, which will be cleaved through Mpro and will result in non-structural proteins and join the RNA to form the virus genome.25 Thus, Mpro inhibition becomes a drug target for the development of therapeutic agents or antiviral drugs against SARS-CoV-2. Researchers are seeking out products with efficient antiviral activity to prevent the SARS-CoV-2 virus replication. The peel of pomegranate (Punica granatum), has substantial studies that encompass antibacterial,26 anti-inflammatory27 and antioxidant28 activities in addition to antiviral activity agaist Influenza virus,29 Human Immunodeficiency virus (HIV)30 and Herpes simplex virus.31 Punicalagin is a phenolic compound present abundantly in pomegranate peel with high molecular weight.32 Punicalagin is found naturally in two anomeric forms α and β differing from the chiral carbon position (Figure 1). Recent studies including α/β anomers33-35 revels that there are differences in the non-covalent interactions profile between these anomers and proteins.
Hence, the present in silico study was aimed to explore the differences in inhibitory potential of α/β anomers of Punicalagin against the main protease viral of SARS-CoV-2 (Mpro).
COMPUTATIONAL DETAILS Preparation of Mpro and α/β anomers The protonation states of the molecules has a significant influence on its mode of interaction with the receptor. Based on the pKa values, the protonation states of the α/β anomers studied were calculated using the ChemAxon's MARVIN software.36 The geometries of the α/β anomers were optimized by using standard techniques.37 Optimization calculations were performed by using Density Functional Theory38 (DFT) method at B3LYP functional39 along with 6-31+G(d,p) basis set implemented in Gaussian 16 package.40 Vibrational modes of the optimized geometries were calculated in order to determine whether the resulting geometries are true minima or transition states (Supplementary Figure 2S). All optimization calculations were performed in solution by using Polarizable Continuum Model (PCM)41 with the Integral Equation Formalism (IEF)42 using water as a solvent. The optimized structures with the lowest energy were used for the molecular docking calculations. The X-ray crystal structure of SARS-Cov-2 main protease (Mpro) in complex with an inhibitor N343 at 2.16 Å resolution (PDB ID: 6LU7) used is this study was obtained from RCSB Protein Data Bank.44 To confirm the quality of the Mpro structural model used in this work, the Ramachandran plot was built using the Ramachandran Plot Server (https://zlab.umassmed.edu/bu/rama/). We can see that the vast majority of the Mpro amino acid residues (99.3%, represented by green crosses) are found in highly permitted regions (black, dark grey, grey and light grey regions) (Supplementary Figure 3S). The Mpro structure was prepared for molecular docking by removing the water molecules and other hetero molecules from the original crystal structure. The molecular docking study Docking Input files were created using AutoDock tools of MGL tools.45,46 The 3D grid box with a box size of 30 Å × 30 Å × 30 Å with the center coordinates x = -13.579, y = 18.940 and z = 69.318, which are the coordinates in the Nε2 of His41 residue that is included in the Mpro catalytic dyad.47 Molecular docking was performed using Autodock Vina.48 Nine best poses were generated for each anomer and scored using Autodock Vina scoring functions. The lowest energy pose of each anomer was used as input for molecular dynamics (DM) simulations. Molecular dynamics simulations All molecular dynamics (MD) simulations were performed using the GROMACS 2018.449 package implemented with the AMBER ff99SB.50 The transferable intramolecular potential with 3 points (TIP3P)51 water molecules were used to solvate the simulated systems. The systems neutralization was achieved through the addition of counter ions. The Leap-Frog algorithm52 was applied to integrate the motion equation with time step of 2.0 fs. The long-range interactions were modeled using particle-mesh Ewald sum (PME)53 with a cut-off of 1.2 nm. The van der Waals interactions were also calculated using the same threshold. Bonds involving hydrogen atoms were restrained using LINCS algorithm.54 The Nosé-Hoover thermostat55 was used to fix the system temperature (310 K) in all production simulations, while the system pressure was controlled using a Parrinello-Rahman barostat56 in the NPT simulations. The geometry of the systems were minimized by the steepest descent algorithm57 for 10000 steps with tolerance of 10 kJ mol-1 nm-1 followed by conjugate gradient algorithm58 for 10000 steps with tolerance of 10 kJ mol-1 nm-1. Two shorts 20-ns equilibrium dynamics with NVT and NPT ensembles were performed. Finally, 200-ns production MD simulation using NVT ensemble was performed in three replicates for each system in order to determine the interaction energy between α/β anomers and Mpro. To analyze the interaction energy between α/β anomers and Mpro we used the Interaction Potential Energy (IPE),59 which can be defined as the total interaction energy between two groups, α/β anomers and Mpro. The IPE calculation includes the sum of van der Waals and electrostatic contributions. To identify non-covalent interactions between anomers and Mpro receptor, we used an open-source protein-ligand interaction profiler (PLIP).60
RESULTS AND DISCUSSION The pKa can predict the protonation states of organic molecules across pH range. Ionizable sites are found often in organic molecules and influence their pharmaceutical properties including target affinity.61 Due to the importance of knowing the protonation state, the pKa values of hydroxyl groups are shown in the Supplementary Figures S1a (α anomer) and S1b (β anomer). We can see that the pKa values do not change with configuration change and then α/β anomers have the same ionizable sites at pH = 7.4, that are indicated by blue arrows in Supplementary Figure 1Sa. Hence, negatively charged punicalagin α/β anomers were considered for this study. In order to improve the structures of α/β anomers their optimized structures were performed by Density Functional Theory (DFT) using level of theory (Figure 2). The vibrational mode values of the α/β anomers were included in the supplementary Figure 2S.
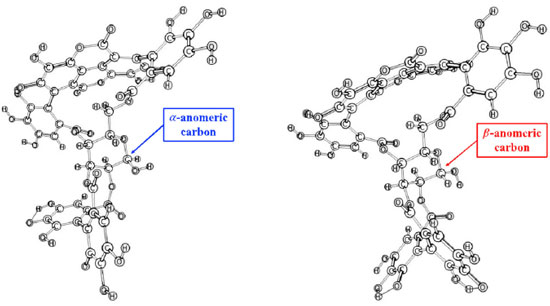 Figure 2. Optimized geometries of α (a) and β (b) anomers by using B3LYP/6 31+G(d,p) level of theory
From α and β optimized structures and using Mpro as receptor, the best poses (pose 1) generated from docking analysis (Supplementary Figure 4S), MD simulations were performed for identifying the α/β anomers intermolecular interaction profile. The interaction potential energy (IPE) calculations were performed to evaluate the affinity as well as the contribution of each residue of binding pocket of Mpro receptor with the ligands. Based on atoms of α/β anomers as reference, the RMSD analysis (Figure 3) showed a structural equilibrium around 125 ns for α (Figure 3A, black, red and green lines) and around 50 ns for β (Figure 3B, blue, orange and purple lines). Thus, the IPE calculations were performed in the last 75 ns for α anomer and 150 ns for the β anomer. The average IPE (standard deviation) between Mpro receptor and α/β anomers were -250.96 kJ mol-1 (20.40) and -320.00 kJ mol-1 (27.05), respectively. Then, according to the IPE values, β anomer has a highest affinity for Mpro compared to the α anomer. This result suggests that the β anomer can become a potential inhibitor for SARS-CoV-2 main protease.
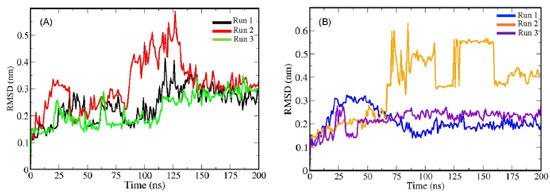 Figure 3. Root Mean Square Deviations (RMSD) for punicalagin α (A; black, red and green lines) and β (B; blue, orange and violet lines) anomers. All MD simulations were performed in three replicates each anomer studied
A closed comparative analysis of the binding of α (Figure 4a) and β (Figure 4b) anomers with pocket Mpro residues in the last simulation frame (200 ns) of MD, revealed that the β anomer binds to the protein through nine favorable hydrogen bonds. On the other hand, a close inspection of the binding of α anomer (Figure 4a) with pocket Mpro residues showed that α anomer ligand binds with the protein with eight hydrogen bonds. Hydrogen bonds contribute from 11 to 60 kJ mol-1 in the interaction potential energy (IPE)62 and are force intermolecular most influential in molecular recognition.63 Furthermore, the β-anomeric carbon configuration (Figure 5) allowed the holding of one hydrogen bond (blue dashed line) and one hydrophobic interaction (gray dashed line) with THR45 Mpro residue. We can see in Figure 5 that one hydrogen bond is formed between hydroxyl groups of β-anomeric carbon of β anomer and residue THR45 at distance of 3.42 Å. In addition, the hydrophobic interaction is formed between Cγ of residue THR45 and C-47 of β anomer. The presence of hydrophobic groups in the ligand can expel molecular water from protein pocket and maximize hydrogen bonds account leading to an increased selectivity and efficiency of the receptor for a certain ligand,64 as in the case of the β-anomer and Mpro receptor, where the β-anomeric configuration of punicalagin allowed access to two more hydrogen bonds and a hydrophobic interaction. In other words, the combination of hydrogen bonds and hydrophobic interaction found between β-anomer and Mpro binding pocket led to higher selectivity and specificity of the β-anomer ligand compared to the α-anomer.
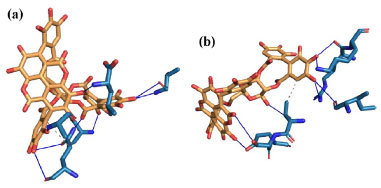 Figure 4. Details of interactions of α- (a) and β- (b) anomers with Mpro receptor residues. blue lines represent hydrogen bonds between α- (a) or β- (b) anomer with pocket Mpro residues
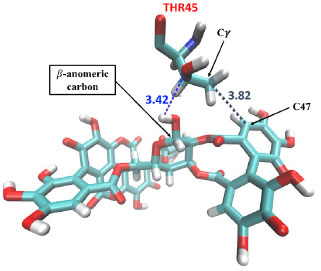 Figure 5. Representative highlight of non-covalent interactions between residue THR45 Mpro and β-anomer. The blue dashed lines are the hydrogen bonds and gray dashed line is the hydrophobic interaction
CONCLUSIONS In this work, we reported a systematic study of affinity and specificity of α- and β-anomers of Punicalagin with Mpro protease of SARS-CoV-2 by using molecular docking and molecular dynamics simulations. Interaction Potential Energy (IPE) calculations through molecular dynamics simulations reveals that the β-anomer of Punicalagin has a higher affinity for the pocket Mpro receptor compared to the α-anomer. Furthermore, β-anomer configuration shows more hydrogen bonds and a hydrophobic interaction than α-anomer with pocket Mpro residues. The amount and combination of hydrogen bonds and hydrophobic interaction can lead to selectivity and specificity of β-anomer of Punicalagin being able to act as a potential drug candidate. Therefore, we way conclude that the β-anomer ligand could act as potential inhibitor against the main protease of SARS-CoV-2. SUPPLEMENTARY MATERIAL Some images of the systems used in this work are available at http://quimicanova.sbq.org.br, in the form of a PDF file, with free access.
ACKNOWLEDGEMENTS The authors thank High-Performance Computing Center (NPAD) at Federal University of Rio Grande do Norte (UFRN), the National High-Performance Processing Center of the Federal University of Ceará (UFC) for providing computational resources and Pró-Reitoria de Extensão of Federal University of Ceará (PROEX/UFC).
REFERENCES 1. Zoumpourlis, V.; Goulielmaki, M.; Rizos, E.; Baliou, S.; Spandidos, D. A.; Mol. Med. Rep. 2020, 22, 3035. [Crossref] 2. Singhal, T.; The Indian Journal of Pediatrics 2020, 87, 281. [Crossref] 3. Salazar, E.; Perez, K. K.; Ashraf, M.; Chen, J.; Castillo, B.; Christensen, P. A.; Eubank, T.; Bernard, D. W.; Eagar, T. N.; Long, S. W.; Subedi, S.; Olsen, R. J.; Leveque, C.; Schwartz, M. R.; Dey, M.; Chavez-East, C.; Rogers, J.; Shehabeldin, A.; Joseph, D.; Williams, G.; Thomas, K.; Masud, F.; Talley, C.; Dlouhy, K. G.; Lopez, B. V.; Hampton, C.; Lavinder, J.; Gollihar, J. D.; Maranhao, A. C.; Ippolito, G. C.; Saavedra, M. O.; Cantu, C. C.; Yerramilli, P.; Pruitt, L.; Musser, J. M.; Am. J. Pathol. 2020, 190, 1680. [Crossref] 4. Suručić, R.; Tubić, B.; Stojiljković, M. P.; Djuric, D. M.; Travar, M.; Grabez, M.; Savikin, K.; Skrbić, R.; Mol. Cell. Biochem. 2021, 476, 1179.b [Crossref] 5. Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D. S. C.; Du, B.; Li, L.; Zeng, G.; Yuen, K.-Y.; Chen, R.; Tang, C.; Wang, T.; Chen, P.; Xiang, J.; Li, S.; Wang, J.; Liang, Z.; Peng, Y.; Wei, L.; Liu, Y.; Hu, Y.; Peng, P.; Wang, J.; Liu, J.; Chen, Z.; Li, G.; Zheng, Z.; Qiu, S.; Luo, J.; Ye, C.; Zhu, S.; Zhong, N.; N. Engl. J. Med. 2020, 382, 1708. [Crossref] 6. He, F.; Deng, Y.; Li, W.; J. Med. Virol. 2020, 92, 719. [Crossref] 7. Berlin, D. A.; Gulick, R. M.; Martinez, F. J.; N. Engl. J. Med. 2020, 383, 2451. [Crossref] 8. Giacomelli, A.; Pezzati, L.; Conti, F.; Bernacchia, D.; Siano, M.; Oreni, L.; Rusconi, S.; Gervasoni, C.; Ridolfo, A. L.; Rizzardini, G.; Antinori, S.; Galli, M.; Clin. Infect. Dis. 2020, 71, 889. [Crossref] 9. Senger, M. R.; Evangelista, T. C. S.; Dantas, R. F.; Santana, M. V. da S.; Gonçalves, L. C. S.; de Souza Neto, L. R.; Ferreira, S. B.; Silva-Junior, F. P.; Mem. Inst. Oswaldo Cruz 2020, 115, 1. [Crossref] 10. Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F.; Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 11727. [Crossref] 11. Zhou, M.; Zhang, X.; Qu, J.; Front. Med. 2020, 14, 126. [Crossref] 12. Fan, C.; Lu, W.; Li, K.; Ding, Y.; Wang, J.; Front. Med. 2021, 7, 1045. [Crossref] 13. Liu, F.; Long, X.; Zhang, B.; Zhang, W.; Chen, X.; Zhang, Z.; Clin. Gastroenterol. Hepatol. 2020, 18, 2128. [Crossref] 14. Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; Guo, Q.; Song, T.; He, J.; Yen, H.-L.; Peiris, M.; Wu, J.; N. Engl. J. Med. 2020, 382, 1177. [Crossref] 15. Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W.; Gut 2020, 69, 1141. [Crossref] 16. Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W.; Am. J. Respir. Crit. Care Med. 2020, 202, 756. [Crossref] 17. Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; Yang, X.; He, L.; Zhang, L.; Yang, Z.; Geng, J.-J.; Chen, R.; Zhang, H.; Wang, B.; Zhu, Y.-M.; Nan, G.; Jiang, J.-L.; Li, L.; Wu, J.; Lin, P.; Huang, W.; Xie, L.; Zheng, Z.-H.; Zhang, K.; Miao, J.-L.; Cui, H.-Y.; Huang, M.; Zhang, J.; Fu, L.; Yang, X.-M.; Zhao, Z.; Sun, S.; Gu, H.; Wang, Z.; Wang, C.-F.; Lu, Y.; Liu, Y.-Y.; Wang, Q.-Y.; Bian, H.; Zhu, P.; Chen, Z.-N.; Signal Transduction Targeted Ther. 2020, 5, 283. [Crossref] 18. Daly, J. L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M. K.; Antón-Plágaro, C.; Shoemark, D. K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; Greber, U. F.; Horvath, P.; Sessions, R. B.; Helenius, A.; Hiscox, J. A.; Teesalu, T.; Matthews, D. A.; Davidson, A. D.; Collins, B. M.; Cullen, P. J.; Yamauchi, Y.; Science 2020, 370, 861. [Crossref] 19. Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L. D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; Smura, T.; Levanov, L.; Szirovicza, L.; Tobi, A.; Kallio-Kokko, H.; Österlund, P.; Joensuu, M.; Meunier, F. A.; Butcher, S. J.; Winkler, M. S.; Mollenhauer, B.; Helenius, A.; Gokce, O.; Teesalu, T.; Hepojoki, J.; Vapalahti, O.; Stadelmann, C.; Balistreri, G.; Simons, M.; Science (1979) 2020, 370, 856. [Crossref] 20. Clausen, T. M.; Sandoval, D. R.; Spliid, C. B.; Pihl, J.; Perrett, H. R.; Painter, C. D.; Narayanan, A.; Majowicz, S. A.; Kwong, E. M.; McVicar, R. N.; Thacker, B. E.; Glass, C. A.; Yang, Z.; Torres, J. L.; Golden, G. J.; Bartels, P. L.; Porell, R. N.; Garretson, A. F.; Laubach, L.; Feldman, J.; Yin, X.; Pu, Y.; Hauser, B. M.; Caradonna, T. M.; Kellman, B. P.; Martino, C.; Gordts, P. L. S. M.; Chanda, S. K.; Schmidt, A. G.; Godula, K.; Leibel, S. L.; Jose, J.; Corbett, K. D.; Ward, A. B.; Carlin, A. F.; Esko, J. D.; Cell 2020, 183, 1043. [Crossref] 21. Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M.; Cell Discovery 2020, 6, 16. [Crossref] 22. Kokic, G.; Hillen, H. S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P.; Nat. Commun. 2021, 12, 279. [Crossref] 23. van Kampen, J. J. A.; van de Vijver, D. A. M. C.; Fraaij, P. L. A.; Haagmans, B. L.; Lamers, M. M.; Okba, N.; van den Akker, J. P. C.; Endeman, H.; Gommers, D. A. M. P. J.; Cornelissen, J. J.; Hoek, R. A. S.; van der Eerden, M. M.; Hesselink, D. A.; Metselaar, H. J.; Verbon, A.; de Steenwinkel, J. E. M.; Aron, G. I.; van Gorp, E. C. M.; van Boheemen, S.; Voermans, J. C.; Boucher, C. A. B.; Molenkamp, R.; Koopmans, M. P. G.; Geurtsvankessel, C.; van der Eijk, A. A.; Nat. Commun. 2021, 12, 267. [Crossref] 24. Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; Duan, Y.; Yu, J.; Wang, L.; Yang, K.; Liu, F.; Jiang, R.; Yang, X.; You, T.; Liu, X.; Yang, X.; Bai, F.; Liu, H.; Liu, X.; Guddat, L. W.; Xu, W.; Xiao, G.; Qin, C.; Shi, Z.; Jiang, H.; Rao, Z.; Yang, H.; Nature 2020, 582, 289. [Crossref] 25. Zhang, R.; Li, Y.; Zhang, A. L.; Wang, Y.; Molina, M. J.; Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 14857. [Crossref] 26. Voravuthikunchai, S. P.; Sririrak, T.; Limsuwan, S.; Supawita, T.; Iida, T.; Honda, T.; J. Health Sci. 2005, 51, 590. [Crossref] 27. Colombo, E.; Sangiovanni, E.; Dell'Agli, M.; Evidence-Based Complementary Altern. Med. 2013, 2013, 1. [Crossref] 28. Oudane, B.; Boudemagh, D.; Bounekhel, M.; Sobhi, W.; Vidal, M.; Broussy, S.; J. Mol. Struct. 2018, 1156, 390. [Crossref] 29. Moradi, M. T.; Karimi, A.; Shahrani, M.; Hashemi, L.; Ghaffari-Goosheh, M. S.; Avicenna J. Med. Biotechnol. 2019, 11, 285. 30. Shaygannia, E.; Bahmani, M.; Zamanzad, B.; Rafieian-Kopaei, M.; J. Evidence-Based Complementary Altern. Med. 2016, 21, 221. [Crossref] 31. Arunkumar, J.; Rajarajan, S.; Microb. Pathog. 2018, 118, 301. [Crossref] 32. Gil, M. I.; Tomás-Barberán, F. A.; Hess-Pierce, B.; Holcroft, D. M.; Kader, A. A.; J. Agric. Food Chem. 2000, 48, 4581. [Crossref] 33. Saparbaev, E.; Aladinskaia, V.; Yamaletdinov, R.; Pereverzev, A. Y.; Boyarkin, O. V.; The J. Phys. Chem. Lett. 2020, 11, 3327. [Crossref] 34. Rozada, T.; de Melo, U.; Pontes, R.; Rittner, R.; Basso, E.; J. Braz. Chem. Soc. 2018, 30, 948. [Crossref] 35. Mazik, M.; Radunz, W.; Sicking, W.; Org. Lett. 2002, 4, 4579. [Crossref] 36. https://chemaxon.com/products/marvin, accessed julho 2022. 37. Chambers, Ll. G.; Fletcher, R.; The Mathematical Gazette 2001, 85, 562. 38. Zinola, C. F. Em Electrocatalysis: Computational, Experimental, and Industrial Aspects; Zinola, C. F., eds.; CRC Press: Boca Raton, 2010. [Crossref] 39. Perdew, J. P.; Burke, K.; Ernzerhof, M.; Phys. Rev. Lett. 1996, 77, 3865. [Crossref] 40. Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Scalmani, G.; Barone, V.; Petersson, G. A.; Nakatsuji, H.; Li, X.; Caricato, M.; Marenich, A. V.; Bloino, J.; Janesko, B. G.; Gomperts, R.; Mennucci, B.; Hratchian, H. P.; Ortiz, J. V.; Izmaylov, A. F.; Sonnenberg, J. L.; Williams-Young, D.; Ding, F.; Lipparini, F.; Egidi, F.; Goings, J.; Peng, B.; Petrone, A.; Henderson, T.; Ranasinghe, D.; Zakrzewski, V. G.; Gao, J.; Rega, N.; Zheng, G.; Liang, W.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Vreven, T.; Throssell, K.; Montgomery, J. A., Jr.; Peralta, J. E.; Ogliaro, F.; Bearpark, M. J.; Heyd, J. J.; Brothers, E. N.; Kudin, K. N.; Staroverov, V. N.; Keith, T. A.; Kobayashi, R.; Normand, J.; Raghavachari, K.; Rendell, A. P.; Burant, J. C.; Iyengar, S. S.; Tomasi, J.; Cossi, M.; Millam, J. M.; Klene, M.; Adamo, C.; Cammi, R.; Ochterski, J. W.; Martin, R. L.; Morokuma, K.; Farkas, O.; Foresman, J. B.; Fox, D. J.; Gaussian 16, Gaussian, Inc., Wallingford CT, 2016. 41. Mennucci, B.; Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012, 2, 386. [Crossref] 42. Mennucci, B.; Cancès, E.; Tomasi, J.; J. Phys. Chem. B 1997, 101, 10506. [Crossref] 43. Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; Duan, Y.; Yu, J.; Wang, L.; Yang, K.; Liu, F.; Jiang, R.; Yang, X.; You, T.; Liu, X.; Yang, X.; Bai, F.; Liu, H.; Liu, X.; Guddat, L. W.; Xu, W.; Xiao, G.; Qin, C.; Shi, Z.; Jiang, H.; Rao, Z.; Yang, H.; Nature 2020, 582, 289. [Crossref] 44. Berman, H. M.; Battistuz, T.; Bhat, T. N.; Bluhm, W. F.; Bourne, P. E.; Burkhardt, K.; Feng, Z.; Gilliland, G. L.; Iype, L.; Jain, S.; Fagan, P.; Marvin, J.; Padilla, D.; Ravichandran, V.; Schneider, B.; Thanki, N.; Weissig, H.; Westbrook, J. D.; Zardecki, C.; Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002, 58, 899. [Crossref] 45. Holt, P. A.; Chaires, J. B.; Trent, J. O.; J. Chem. Inf. Model. 2008, 48, 1602. [Crossref] 46. Morris, G. M.; Ruth, H.; Lindstrom, W.; Sanner, M. F.; Belew, R. K.; Goodsell, D. S.; Olson, A. J.; J. Comput. Chem. 2009, 30, 2785. [Crossref] 47. Chhetri, A.; Chettri, S.; Rai, P.; Mishra, D. K.; Sinha, B.; Brahman, D.; J. Mol. Struct. 2021, 1225, 1. [Crossref] 48. Trott, O.; Olson, A. J.; J. Comput. Chem. 2009, 31, 1. [Crossref] 49. Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A. E.; Berendsen, H. J. C.; J. Comput. Chem. 2005, 26, 1701. [Crossref] 50. Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J. L.; Dror, R. O.; Shaw, D. E.; Proteins: Struct., Funct., Bioinf. 2010, 78, 1950. [Crossref] 51. Jorgensen, W. L.; Chandrasekhar, J.; Madura, J. D.; Impey, R. W.; Klein, M. L.; J. Chem. Phys. 1983, 79, 926. [Crossref] 52. Van Gunsteren, W. F.; Berendsen, H. J. C.; Mol. Simul. 1988, 1, 173. [Crossref] 53. Darden, T.; York, D.; Pedersen, L.; J. Chem. Phys. 1993, 98, 10089. [Crossref] 54. Hess, B.; Bekker, H.; Berendsen, H. J. C.; Fraaije, J. G. E. M.; J. Comput. Chem. 1997, 18, 1463. [Crossref] 55. Hoover, W. G.; Phys. Rev. A 1985, 31, 1695. [Crossref] 56. Nosé, S.; Klein, M. L.; Molecular Physics 1983, 50, 1055. [Crossref] 57. Arfken, G. B.; Weber, H. J.; Harris, F. E.; Mathematical Methods for Physicists: A Comprehensive Guide, 7th ed., Academic Press: Waltham, 2013. 58. Hestenes, M. R.; Stiefel, E.; J. Res. Natl. Bur. Stand. 1952, 49, 409. [Crossref] 59. Amorim-Carmo, B.; Daniele-Silva, A.; Parente, A. M. S.; Furtado, A. A.; Carvalho, E.; Oliveira, J. W. F.; Santos, E. C. G.; Silva, M. S.; Silva, S. R. B.; Silva-Júnior, A. A.; Monteiro, N. K.; Fernandes-Pedrosa, M. F.; Int. J. Mol. Sci. 2019, 20, 623. [Crossref] 60. Salentin, S.; Schreiber, S.; Haupt, V. J.; Adasme, M. F.; Schroeder, M.; Nucleic Acids Res. 2015, 43, 443. [Crossref] 61. Işık, M.; Rustenburg, A. S.; Rizzi, A.; Gunner, M. R.; Mobley, D. L.; Chodera, J. D.; J. Comput.-Aided Mol. Des. 2021, 35, 131. [Crossref] 62. Pace, C. N.; Fu, H.; Fryar, K. L.; Landua, J.; Trevino, S. R.; Schell, D.; Thurlkill, R. L.; Imura, S.; Scholtz, J. M.; Gajiwala, K.; Sevcik, J.; Urbanikova, L.; Myers, J. K.; Takano, K.; Hebert, E. J.; Shirley, B. A.; Grimsley, G. R.; Protein Sci. 2014, 23, 652. [Crossref] 63. Dong, J.; Davis, A. P.; Angew. Chem., Int. Ed. 2021, 60, 8035. [Crossref] 64. Yao, H.; Ke, H.; Zhang, X.; Pan, S.-J.; Li, M.-S.; Yang, L.-P.; Schreckenbach, G.; Jiang, W.; J. Am. Chem. Soc. 2018, 140, 13466. [Crossref] |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access