Nota Técnica
| Rapid method for simultaneous determination of cis-9, trans-11 and trans-10, cis-12 conjugated linoleic acid isomers in milk by GC-FID |
|
Brenda Lee Simas PortoI,II; Isaura Daniele Leite FariaI; Jéssica Cordeiro Queiroz de SouzaI; Olívia Brito de Oliveira MoreiraI; Marco Antônio Sundfeld da GamaIII; Marcone Augusto Leal de OliveiraI,IV,*
I. Departamento de Química, Instituto de Ciências Exatas, Universidade Federal de Juiz de Fora, 36036-330 Juiz de Fora − MG, Brasil Recebido em 31/08/2022 *e-mail: marcone.oliveira@ufjf.br Ruminant milk is the lead source of conjugated linoleic acid (CLA) in the human diet, with cis-9, trans-11 CLA being the major among all. Small amounts of trans-10, cis-12 CLA are also found in synthetic supplements. Since both isomers are biologically active with potential health benefits, there is great interest in quantifying them for quality control routines. An alternative method for the analysis of the aforementioned CLAs by fast gas chromatography (GC) is discussed in the present study. The fatty acid methyl ester mixture obtained by alkaline catalysis was injected into a GC equipped with a flame ionization detector (FID) and fitted with an ionic liquid SLB-IL111 chromatographic column (15 m × 0.10 mm × 0.08 µm). Separation was achieved in less than 5 min using a 168 ºC isotherm run. Both CLA isomers were quantified by using of single point standard addition statistical approach. Results were contested to those obtained using the recommended 100 m long CP-Sil88 capillary column and none evidence of significant differences was found within 95% confidence interval. Therefore, the proposed method could be valuable to focused regulatory routines of large numbers of samples with greater analytical frequency. INTRODUCTION Conjugated linoleic acid (CLA) is a group of positional and geometric isomers of linoleic acid (C18:2 n-6) distinguished by conjugated double bonds, mainly found in dairies produced with milk from ruminants. Although more than 20 CLA isomers were identified in ruminant milk-containing foods, two of them (cis-9, trans-11 and trans-10, cis-12) are particularly interesting due to their potential health benefits observed in several related studies conducted in the last three decades.1-3 Dairy fat is the lead source of CLA in the human diet, with cis-9, trans-11 CLA being the predominant isomer. Additionally, even though trans-10, cis-12 CLA usually represents less than 1% of total CLA in milk fat, it is present at higher concentrations in most synthetic CLA supplements.4,5 Therefore, considering the biochemical relevance of both CLA isomers, there is relevant interest in quantifying them in milk derivative foods and CLA supplements for quality control and adulteration traceability purposes. Gas chromatography with flame ionization detection (GC-FID) is the official technique used for the analysis of fatty acids (FA) composition in foods (AOAC 996.06).6 High polar capillary columns coated with cyanopropyl siloxane such as SP-2560 or CP-Sil88 (100 m or longer) are usually required for the analysis of CLA isomers within complex matrixes such as fat-containing foods.7 Although these columns enable good peak resolution for most CLA isomers, long running times (~80 minutes) are needed under typical GC-FID operating conditions,8 which makes the analysis very time consuming. Recently, other columns with variable lengths and highly polar stationary phases (SP) have been made available in supplier companies to improve the separation of CLA isomers while reducing the total running time.9-11 In the same period, several studies have compared the efficiency of the new GC columns with those recommended for separating critical FA, such as fatty acids methyl esters (FAME), in different foods, including milk fat.7,12-19 Ionic liquid (IL) based GC columns have already been considered for separating critical CLA isomers in milk fat, such as cis-9, trans-11, and trans-7, cis-9 CLA, which coelute in the most recommended CP-Sil88 and SP-2560 columns. However, 100 m and 200 m long IL columns were needed, which resulted in long running times despite achieving a resolution ≥ 1.5.7,13 The cis-9, trans-11, and trans-7, cis-9 CLA isomers in ruminant tissue were separated in 12.5 min using an SLB-IL111 column with 30 m by Turner et al.9 An SLB-IL111 column with a 15 m length was previously used for biodiesel analysis,11,12,16,18 however, to our knowledge, it has not been applied for fatty acid analysis in milk samples yet. Different from the method described by Turner et al.,9 which uses a longer column and focuses on separating other CLA isomers, in the present work, a rapid method by GC-FID was developed and optimized, in order to separate and quantify the cis-9, trans-11 and trans-10, cis-12 CLA isomers in raw milk by using a 15 m long SLB-IL111 column. The proposed method could be an advantageous alternative for further analysis focused on assessing the CLA content of large groups of samples for regulatory purposes with enhanced analytical throughput.
EXPERIMENTAL Reagents and solutions The analytical grade reagents and solvents, such as methanol, hexane, isopropanol, acetic acid, anhydrous sodium sulfate, sodium hydroxide, sodium methoxide solution and the FAME mix were all purchased from Sigma-Aldrich (St Louis, MO, USA). The standards mixture from BASF (BA, Brazil), containing methyl palmitate (C16:0 ME), methyl stearate (C18:0 ME), methyl linoleate (C18:2 ME), and CLA isomers (cis-9, trans-11 and trans-10, cis-12) were kindly provided by EMBRAPA (MG, Brazil). Water was purified by deionization using the Milli-Q system with a resistivity of 18.5 MΩ cm. Individual solutions of FAME were prepared by dissolving appropriate amounts in hexane at a concentration of 1.0 mmol L-1. All solutions were stored in a freezer at -4 ºC until analysis. GC-FID instrumentation The analysis of the FAME was performed on a GC-2010 Plus gas chromatograph from Shimadzu (Kyoto, Japan), with auto-injector AOC-i-20 and detection by flame ionization (FID). Two fused silica capillary columns were used, one with polar stationary phase (CP-Sil88 for FAME; 100 m × 0.25 mm × 0.2 µm) from Agilent Technologies (Palo Alto, CA, USA) and another with ionic liquid (SLB-IL111; 15 m × 0.10 mm × 0.08 µm) from Supelco (Bellefonte, USA). The chromatographic conditions for CP-Sil88 column were described by Cruz-Hernandez et al.8 For the SLB-IL111 column, the analytical conditions were adapted from Delmonte et al.7 and set as follows: injection volume of 1.0 µL in split mode with a ratio of 1:100; injector and detector to 250 ºC; flow rate of carrier gas (H2) of 1.0 mL min-1; linear velocity of 26 cm s-1 and isotherm at 168 ºC. The compounds were identified by co-injection of standards and by comparison of the retention time of a standard mixture. The FAME composition was determined by the standard addition of the individual's standard solutions and are expressed in g L-1. Samples preparation The samples of raw milk were kindly provided by EMBRAPA. The samples were submitted to the lipid extraction and transesterification procedures. In the lipid extraction procedure was used hexane and isopropanol as the organic phase.20 The solvents were evaporated in nitrogen flow until only the lipid phase remained in the test tube. Samples were prepared and analyzed in duplicate. For alkaline transesterification catalysts, sodium methoxide in methanol was used. The samples were transferred and stored in a capped vial in a freezer at -20 ºC until analysis.21 Procedure for quantification Quantification by single point standard addition (SPSA) method, based on equations (1) and (2), was performed in the trials with both chromatographic columns specified in the section GC-FID instrumentation. This approach is suitable for milk samples because it considers the matrix effects and uses only two injections to determine analyte concentration.22,23 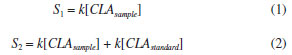 S1 is the analyte signal within the sample chromatogram; k is a constant of proportionality; [CLAsample] is the concentration of analyte in the sample; S2 is the analyte signal in the solution containing both sample and standard, and [CLAstandard] is the concentration of the added standard in the sample. Two solutions were prepared, the first one containing only the extracted sample (920 µL of sample + 80 µL of hexane) and the second containing the extracted sample and standard (80 µL of standard + 920 µL of the sample). By isolating k and equating equations (1) and (2) we have equation (3): 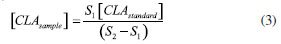 when the concentration of CLA standard [CLAstandard] is 2.7 g L-1 and the volumes used in the dilution are the above volumes, the equation 3 can be rearranged into equation 4, used to calculate the concentration of the CLA in each sample [CLAsample]: 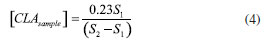
RESULTS AND DISCUSSION Figure 1(A) shows the resulting chromatogram of a CLA standard mixture together with a FAME mix analyzed in a 100 m long CP-Sil88 column with a programmed heating ramp.8 Since traditional FAME mix do not include trans-FA, CLA provides complementary information for full FA profile identification considering milk derivative analysis, therefore, from this analysis, the composition compiling majoritarian FAME and CLA isomers was established. The final mixture was then used as a secondary standard for all subsequent measurements.
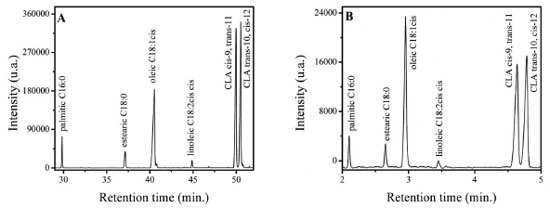 Figure 1. Chromatogram of isomers of CLA and FAME mix resulting from the application of the recommended method with column CP-Sil88 (A) and the alternative method with SLB-IL111 (B). CLA standards at a concentration of 2.52 g L-1. Experimental conditions: (A) Column CP-Sil88 (100 m × 0.25 mm × 0.2 µm); injection: 250 ºC, 1.0 µL, in split mode 1:50; detector: 250 ºC; carrier gas: H2 at flow rate of 1.0 mL min-1; temperature programming: 45 ºC by 4 min, a linear increase of 13 ºC min-1 up to 175 ºC, remaining 175 ºC by 27 min, a linear increase of 4 ºC min-1 up to 215 ºC, and remaining at 215 ºC by 35 min. (B) SLB-IL111 column (15 m × 0.1 mm × 0.08 µm); injection: 250 ºC, 1.0 µL, in split mode 1:100; detector: 250 ºC; carrier gas: H2 at flow rate of 1.0 mL min-1; temperature programming: 45 °C by 4 min, a linear increase of 13 °C min-1 up to 175 °C, remaining 175 °C by 27 min, a linear increase of 4 °C min-1 up to 215 °C, and remaining at 215 °C by 35 min. (B) SLB-IL111 column (15 m × 0.1 mm × 0.08 μm); injection: 250 °C, 1.0 μL, in split mode 1:100; detector: 250 °C; carrier gas: H2 at flow rate of 1.0 mL min-1; temperature programming: isotherm of 168 °C
Furthermore, it is not possible to measure by area normalization in this case because some small FAME can coelute with the solvent peak in the proposed method, as will be discussed forward. Figure 1(B) shows the same CLA-FAME mixture analyzed by the proposed method using SLB-IL111 column with 15 m in length and isotherm at 168 ºC. The separation profile is maintained for both analyzes, however, the time spent in the first analysis is about 10 times greater than the second column to separate the isomers of CLA. To evaluate if the FAME-CLA profile would be found in real samples, a milk sample prepared according to the optimized protocol described in the section Samples preparation was injected using both columns (Figures 2(A) and 2(B)). The separation profile of the standard mixture was replicated in the analysis of milk. To finally confirm the identification of the CLA isomer present, the sample was spiked with CLA standards (Figures 2(C) and 2(D)). By comparison of the retention times shown in Figure 1, the CLA isomer within the sample is the cis-9, trans-11. It is noticeable that the proposed method was selective for the rapid separation of the isomers of CLA of interest.
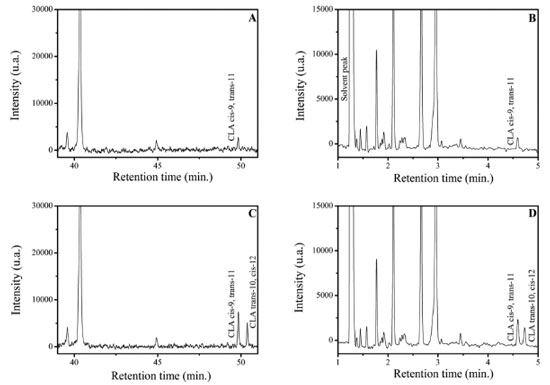 Figure 2. Chromatograms of milk samples with and without of CLA isomers cis-9, trans-11 and trans-10, cis-12 spike. Chromatograms 2(A) and 2(C) resulting from analysis with CP-Sil88 column and chromatograms 2(B) and 2(D) resulting from analysis in column SLB-IL111. Experimental conditions according to described in Figure 1
The method proved to be selective for the CLA isomers cis-9, trans-11 and trans-10, cis-12, however, it still presents a coelution region between 1.0 and 3.5 minutes of analysis. Some peaks coelute, including with the solvent peak, as can be seen in Figure 2(B), which made us rule out the possibility of quantification by area normalization.24 The compounds C15:0, C19:0 and C19:1 were tested as internal standards (IS), but they all eluted in the coelution region and could not be used as IS. Therefore, the quantification by a response factor was also discarded. Thus, the SPSA method was found to be more suitable for the quantification of CLA cis-9, trans-11 isomers in milk samples. The average values and the respective standard deviations (SD) for the quantitative determination of CLA in two milk samples analyzed in duplicate are shown in Table 1.
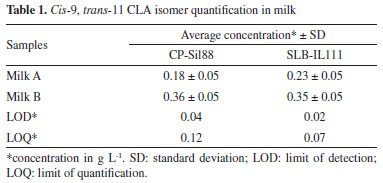
After confirming the normality assumption (p-value > 0.05) in observations, the paired t-test was performed to compare the determinations of both methods. The p-value obtained was 0.14, so that concludes that at a level of 95% confidence do not have statistical differences between these two approaches to quantify CLA isomer cis-9, trans-11 content in milk samples.
CONCLUSIONS By using a column with different SP, it was possible to separate and quantify the CLA isomer cis-9, and trans-11 in milk samples. The main contribution of this proposed alternative method of CLA is the enhanced throughput output when compared to a recommended method due to the reduction of analysis time to obtain the results along with the reduced gas spent. Finally, the proposition made in this work is very compelling and useful to be considered in analytical routines approaching CLA isomer monitoring in laboratories dedicated to this type of analysis.
ACKNOWLEDGEMENTS The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-475055/2011-0, 301689/2011-3, 424032/2018-0, 303867/2020-5); Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG-CEX-PPM 00398-13, APQ-00585-21); Rede Mineira de Química (CEX.RED- 00010-14); Financiadora de Estudos e Projetos (FINEP) (CT-INFRA 01/2013-REF 0633/13); Instituto Nacional de Ciência e Tecnologia Bioanalítica (INCTBio) (FAPESP 2014/50867-3, CNPq 465389/2014-7, 401256/2020-0) for financial support.
REFERENCES 1. Chilliard, Y.; Glasser, F.; Ferlay, A.; Bernard, L.; Rouel, J.; Doreau, M.; Eur. J. Lipid Sci. Technol. 2007, 109, 828. [Crossref] 2. Koba, K.; Yanagita, T.; Obes. Res. Clin. Pract. 2014, 8, e525. [Crossref] 3. Pariza, M. W.; Park, Y.; Cook, M. E.; Prog. Lipid Res. 2001, 40, 283. [Crossref] 4. McCrorie, T. A.; Keaveney, E. M.; Wallace, J. M. W.; Binns, N.; Livingstone, M. B. E.; Nutr. Res. Rev. 2011, 24, 206. [Crossref] 5. Yu, L.; Adams, D.; Watkins, B. A.; J. Food Compos. Anal. 2003, 16, 419. [Crossref] 6. AOAC International; AOAC 996.06 - Fat (Total, Saturated, and Unsaturated) in Foods, 2010. [Link] accessed in February 2023 7. Delmonte, P.; Fardin Kia, A. R.; Kramer, J. K. G.; Mossoba, M. M.; Sidisky, L.; Rader, J. I.; J. Chromatogr. A 2011, 1218, 545. [Crossref] 8. Cruz-Hernandez, C.; Kramer, J. K. G.; Kennelly, J. J.; Glimm, D. R.; Sorensen, B. M.; Okine, E. K.; Goonewardene, L. A.; Weselake, R. J.; J. Dairy Sci. 2007, 90, 3786. [Crossref] 9. Turner, T.; Rolland, D. C.; Aldai, N.; Dugan, M. E. R.; Can. J. Anim. Sci. 2011, 91, 711. [Crossref] 10. Anderson, J. L.; Ding, J.; Welton, T.; Armstrong, D. W.; J. Am. Chem. Soc. 2002, 124, 14247. [Crossref] 11. Merck; Ionic Liquid GC Capillary Columns, available at https://www.sigmaaldrich.com/BR/pt/technical-documents/protocol/analytical-chemistry/gas-chromatography/ionic-liquid-gc-capillary-columns, accessed in February 2023. 12. Buchanan, M. D.; Stenerson, K. K.; Sidisky, L. M.; SLB-IL111 for Fatty Acid Methyl Ester (FAME) Applications, available at https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/marketing/global/documents/189/128/t411139.pdf, accessed in February 2023. 13. Delmonte, P.; Fardin-Kia, A. R.; Kramer, J. K. G.; Mossoba, M. M.; Sidisky, L.; Tyburczy, C.; Rader, J. I.; J. Chromatogr. A 2012, 1233, 137. [Crossref] 14. Delmonte, P.; Hu, Q.; Kia, A. R. F.; Rader, J. I.; J. Chromatogr. A 2008, 1214, 30. [Crossref] 15. Payagala, T.; Zhang, Y.; Wanigasekara, E.; Huang, K.; Breitbach, Z. S.; Sharma, P. S.; Sidisky, L. M.; Armstrong, D. W.; Anal. Chem. 2009, 81, 160. [Crossref] 16. Ragonese, C.; Tranchida, P. Q.; Sciarrone, D.; Mondello, L.; J. Chromatogr. A 2009, 1216, 8992. [Crossref] 17. Ragonese, C.; Tranchida, P. Q.; Dugo, P.; Dugo, G.; Sidisky, L. M.; Robillard, M. V.; Mondello, L.; Anal. Chem. 2009, 81, 5561. [Crossref] 18. Sato, R. T.; Stroppa, P. H. F.; da Silva, A. D.; de Oliveira, M. A. L.; Quim. Nova 2015, 39, 3. [Crossref] 19. Moltó-Puigmartí, C.; Castellote, A. I.; López-Sabater, M. C.; Anal. Chim. Acta 2007, 602, 122. [Crossref] 20. Hara, A.; Radin, N. S.; Anal. Biochem. 1978, 90, 420. [Crossref] 21. Kuksis, A. In Advances in Lipid Methodology - Two; Christie, W. W., ed.; The Oily Press: Dundee, 1993. [Crossref] 22. Cardone, M. J.; Palermo, P. J.; Sybrandt, L. B.; Anal. Chem. 1980, 52, 1187. [Crossref] 23. Neves, L. N. O.; de Oliveira, M. A. L.; LWT--Food Sci. Technol. 2020, 118, 108766. [Crossref] 24. Official Methods Recommended Practices of the AOAC; AOAC Ce 1f-96 Surplus,1996. [Link] accessed in February 2023 |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access





