Artigo
| Schinopsis brasiliensis bark metabolomic profiling based on NMR: evaluation of metabolic variations of different microenvironment and seasonal periods |
|
Licia dos Reis LuzI,II; Elenilson Godoy Alves FilhoIII; Maria Madalena da Silva SoaresIV; Diogo Denardi PortoV; Helena BeckerI; Guilherme Julião ZocoloII,*
I. Departamento de Química Analítica e Físico-Química, Universidade Federal do Ceará, 60455-760 Fortaleza - CE, Brasil Recebido em 20/02/2023 *e-mail: guilherme.zocolo@embrapa.br Schinopsis brasiliensis Engl. (braúna) is a tree species of the Anacardiaceae family. It is native to Brazil, Paraguay, and Bolivia. It is a candidate for agro-industrial system that can be used to produce phytotherapeutics and bioproducts. However, to comprehensively understand the characteristics of plant within the framework of sustainable development for agro-industries, exploring the chemical and phenotypic diversity of samples from various microenvironments across different collection periods is essential. Accordingly, herein, the influence of different environmental factors and sample collection durations on braúna samples was investigated over several years using multivariate statistical analyses associated with nuclear magnetic resonance (NMR) spectroscopy. Bark samples from different areas and environmental data from different locations were analyzed. Distinct chemical profiles were observed across reserve, forest, and management areas, all marked by elevated sugar levels suggesting water stress adaptation. High radiation and low rainfall correlated with increased sugar and amino acids. Each microenvironment responded uniquely to these environmental conditions. Consequently, plant samples from different microenvironments collected over different periods were observed to respond differently to environmental factors, such as solar radiation and temperature. We correlated the metabolomic data with climatic data evaluating the influence of abiotic factors on the production of metabolites in braúna bark samples. INTRODUCTION Schinopsis brasiliensis Engl. , commonly known as baraúna or braúna, is a tree species native to Brazil, Paraguay, and Bolivia. This tree is highly valued for its excellent wood quality, strength, and long-term durability. It is a versatile tree species with a wide range of applications, particularly in the therapeutic field, wherein it is used to treat various ailments such as influenza, osteoporosis, wounds, diarrhea, and fungal infections. It is also known for its antiseptic and anti-inflammatory properties.1-5 Additionally, it is used as an ornamental plant in urban landscaping projects. Moreover, it is used during central bed afforestation and for the development of parks. Its high-quality wood is utilized as firewood, stakes, and charcoal, and its bark is a valuable source of tannins used in the tannery industry.6 Furthermore, the plant provides sustenance to goats and sheep, contributing to their nutritional value. S. brasiliensis is native to the Caatinga dry forest and thrives in regions marked by low precipitation levels, uneven rainfall distributions, high evapotranspiration rates, and poor water retention capacity of predominantly shallow and rocky soils. Consequently, S. brasiliensis is simultaneously and persistently exposed to numerous environmental stressors.7 Generally, various environmental factors influence the amounts and compositions of metabolites biosynthesized by plants. Biotic and abiotic factors, such as temperature, radiation, infections, and herbivory, induce defense mechanisms that trigger complex biochemical processes altering the chemical compositions of metabolites produced by the affected plants.8,9 Thus, noting that these primary metabolites are important precursors of specialized metabolites and that changes in one type of metabolism often influence another is important.8,9 Photosynthesis is among the most sensitive mechanisms affected by abiotic stress. Plants can be significantly affected when exposed to conditions such as saline, hot, cold, drought, and carbon assimilation. Moreover, primary metabolites can be significantly affected when exposed to these conditions.10,11 The concentrations of metabolites such as sugars, amino acids, and sugar alcohols in plant tissues are most affected by stresses generated by the impairment of CO2 assimilation and complex regulatory networks.10,12 Typically, the biosynthesis of primary and specialized metabolites involves the production of different intermediates under conditions of the same metabolic pathways. Different substrates, energies, and cofactors for the production of secondary enzymes are generated under the action of the primary enzymes. In addition to biotic and abiotic factors, the rate of plant growth and productivity parameters such as its height, crown diameter, and diameter at ground level are directly metabolic and environmental conditions.13 Therefore, members of the same species may grow at different rates depending on the environmental conditions prevalent in that region.14,15 Analytical techniques such as nuclear magnetic resonance (NMR) and mass spectrometry are widely used in metabolomic studies. Among these, NMR spectroscopy is a simple, robust, and comprehensive technique that can be used to analyze small amounts of samples, which can be reused after their analysis.16 The use of the NMR technique adds value to metabolomics as it complements mass spectrometry and plays an important role in fingerprinting and characterizing metabolomic profiles of plants.17 Notably, a combination of chemometric analysis and NMR techniques can be used to qualitatively analyze a large volume of data and realize high throughput screening within short times.18 The aim of this study was to assess chemical variability in the bark samples of S. brasiliensis collected periodically over three years from different fields. The samples were chemically characterized using a combination of NMR spectroscopy and chemometric analysis techniques. The obtained data were correlated with environmental variables, such as temperature, radiation, precipitation, relative humidity, and reference evapotranspiration, and dendrometric parameters of the analyzed samples, such as crown diameter, diameter at ground level, and plant height. The underlying objective was to draw parallels between these sources of variation and the chemical compositions of the samples to identify the property exerting the most significant influence on the metabolome of S. brasiliensis bark samples.
EXPERIMENTAL PART Plant material S. brasiliensis bark samples were collected from three experimental areas in Petrolina and Lagoa Grande (Pernambuco, Brazil). Notably, one of these areas is a preserved area (9º 05' 69.69'' S; 40º 31' 57.88'' W) with native vegetation of the Caatinga dry forest, and it is unaffected by animal grazing. Members of S. brasiliensis can be found in this region in conditions that are comparable to natural ones. The second region is a partially preserved area known as a management area (9º 06' 06.22'' S; 40º 31' 08.34'' W). This region, however, experienced moderate levels of disturbances owing to thinning activities for experimental purposes conducted in 2007.19 The third region is a forest (devoid of shrubs and arboreal strata and consists of rows of plants of certain species, including aroeira and braúna) established in 1979.20 The plant material was sampled four times every year, and the bark of S. brasiliensis was collected. The collection processes were executed over three consecutive years (2015-2018) to represent seasonal variations experienced by the natural populations of the plant. The irregular nature of the semi-arid climate was considered during the analytical process. In total, seven collection cycles were conducted. For each collection cycle, 10 plant samples were collected from each area, amounting to 70 bark samples collected from the management, forest, and reserve sites. Overall, 210 samples of S. brasiliensis bark were collected. Voucher specimens were deposited in the "Herbário do Trópico Semiárido" herbarium (Petrolina, PE, Brazil) under the number HTSA7691. All samples and voucher specimens were prepared following standard operating procedures. Access to genetic resources was registered with the Genetic Heritage Management Council (Conselho de Gestão do Patrimônio Genético (CGEN)) under code A579378, following the Brazilian implementation of the Nagoya Protocol.21 After collection, the samples were dried for a week in a circulating air oven operated at 60 ºC and stored in the dark in falcon tubes (15 mL) for further analysis. The dried plant material was ground using a ball mill. The samples were conditioned in closed transparent plastic bags; adequately labeled; and protected against exposure to excess heat, humidity, and light. Climate data Climatic data of the collection areas were obtained from the database of the Agrometeorological Station of Campo Experimental da Caatinga, Petrolina, PE. The geographic coordinates of these areas are as follows: latitude: 09º13' S; longitude: 40º29' W.22 Various environmental variables were used for the analysis (average temperature, humidity, radiation, precipitation, wind speed and reference evapotranspiration). The data were obtained from the beginning of 2016 to the end of 2018, spanning a period of two years. Graphs for each variable can be found in the supplementary material files. Preparation of extracts for the NMR-spectroscopy-based analysis The samples were prepared using 50.0 mg of bark samples collected from different areas. The samples were ground and suspended in a deuterated solvent (700 μL D2O, Sigma-Aldrich, purity: 99.8%). The extracts were placed in an ultrasonic bath for 5 min. The undissolved extract was separated from the solution via centrifugation (time: 10 min). Finally, a Pasteur pipette was used to filter and transfer the extract to NMR tubes (5 mm). Analysis of the metabolic profile using NMR spectroscopy The acquisition and processing parameters adopted for the recording of NMR spectra were determined based on a previously reported methodology.23 The NMR spectra were recorded using a 600 MHz Agilent DD2 instrument (for 1H core) equipped with a One Probe (1H-19F/15N-31P; internal diameter: 5 mm) for inverse detection. The field gradient was directed along the Z-axis, and the experiments were conducted in the Laboratório Multiusuário de Química de Produtos Naturais (LMQPN) of Embrapa Agroindústria Tropical, in Fortaleza, CE, Brazil. The components were identified by analyzing the spectral profiles recorded using two-dimensional (2D) NMR techniques such as gradient correlation spectroscopy, gradient heteronuclear single quantum coherence, and gradient heteronuclear multiple bond correlation. Additional details regarding the molecular structures, 1H and 13C chemical shifts, multiplicities, correlations, coupling constants, and the methods followed for 2D NMR data acquisition and processing can be found in the supplementary material. The data were analyzed using the data presented in an open-access database (www.hmdb.ca) and by referring to literature reports.24-26 The 1H NMR spectra were recorded in triplicate using the PRESAT pulse sequence (Agilent pulse program library) for water suppression at 4.82 ppm (-1 dB, 41 Hz). The acquisition parameters were as follows: acquisition time, 5.0 s; relaxation delay, 25.0 s; number of scans, 48 (transients); spectral window, 16.0 ppm; fixed temperature, 298 K. The TMSP-d4 signal was used as the internal standard (0.0 ppm). Multivariate statistical analysis of the NMR dataset Different multivariate statistical approaches were adopted to analyze the 1H NMR dataset obtained for different aqueous bark extracts obtained from S. brasiliensis. The spectral region between δ 0.5 and 9.0 was selected for our analysis, and a numerical matrix with a dimensionality of 505,890 data points (63 spectra × 8,030 variables in each spectrum) was generated. The spectral data were converted to American Standard Code for Information Interchange files for matrix construction and were imported using the Origin™ 9.4 program.27 Following this, the data were exported for the multivariate statistical analysis. This data analysis was conducted using Matlab™28 and PLS Toolbox™29. Baseline correction was realized using algorithms. Variable alignment was performed using the correlation-optimized warping technique. A segment of 50 data points and a slack of five data points were used to obtain the results. Normalization techniques were also used to analyze the variables. The data were mean-centered, and the Simplified PLS algorithm was used to decompose the matrix for classification evaluation using the partial least squares discriminant analysis (PLS-DA) technique.30 The process of multivariate statistical analysis was divided into three main parts. Chemometric analysis - part 1: evaluation of the effects of different microenvironments on the metabolic profile of S. brasiliensis The PLS-DA technique was used for this analysis, and the mean values of the response intensities corresponding to each identified metabolite were recorded. The data were recorded over three years during the collection of S. brasiliensis bark samples from different microenvironments. Notably, the effects of the collection period were not considered, and our aim was to study the differences between the expression levels of metabolites across different microenvironments (forest, management, and reserve). The average of the signal intensities of each identified metabolite in the respective microenvironments (forest, management, and reserve) was determined based on the data collected over three years. The results revealed differences in metabolic expression levels between the three microenvironments. Chemometric analysis - part 2: evaluation of dendrometric parameters in different microenvironments The matrix used to describe the previously reported dataset was adjusted to be compatible with and correlate with important dendrometric variables related to growth conditions; this was done to correlate the bark composition (metabolic profile) of S. brasiliensis with growth conditions (mean environmental temperature, wind speed, reference evapotranspiration, tree height, diameter at ground level, biomass, relative humidity of air, incident solar radiation amount, and degree of precipitation). As stated, the PLS-based multivariate regression analysis was performed using various autoscaled values as categorical variables (Y matrix).31 The number of latent variables (LV) for both classification and regression modeling (PLS-DA and PLS) was selected based on different statistical parameters: root mean square error of calibration (RMSEC), root mean square error of cross-validation (RMSECV), and the respective correlation coefficients (r2). The Venetian blind method (five splits and one sample per split) was used for analysis during cross-validation.30 Chemometric analysis - part 3: correlation of environmental factors and collection periods The third component of the chemometric analysis involved the correlation of the collection period and environmental variables (temperature, wind speed, reference evapotranspiration, relative humidity, incident solar radiation, and precipitation) with the metabolites in the bark of S. brasiliensis. The spectral region analyzed in Part 1 was used for this analysis. Multivariate statistical analyses were performed using R 4.0.332 and SIMCA33 14.1. The environmental data were transformed based on the data type before executing the analytical processes. The relative air humidity was transformed using the arcsine method for data expressed as percentages, while the other data were converted to a logarithmic scale.14 A canonical correlation analysis (CCA) was performed using the vegan R package to correlate environmental data and collection periods with the metabolites in the samples. The orthogonal partial least squares discriminant analysis (OPLS-DA), a supervised method, was implemented using the same dataset (constructed using only NMR data) to identify the discriminating variables that could be used to differentiate the groups identified based on the PCA and CCA. 1H Quantitative (q)NMR-spectroscopy-based analysis Bioactive compounds were quantified following an external reference method. A standard sucrose solution (5.0 mg L-1) was used to calibrate the spectrometer, and the probe was updated with the necessary concentration determination parameters.34 The signals at δ 5.43 (sucrose), δ 5.25 (α-glucose), δ 4.67 (β-glucose), δ 4.14 (fructose), δ 7.01 (gallic acid), and δ 2.03 (acetic acid) were selected for quantification, and these selections were made based on the signal intensities. The combined uncertainty of the method was estimated based on the analytical errors and standard deviations of the three sampling repetitions. The one-way analysis of variance (ANOVA) technique was used to evaluate the results at a significance level of 0.05. The mean values were compared based on Tukey's tests to statistically assess differences between concentrations, and Levene's test was conducted to test for homogeneity of variance.
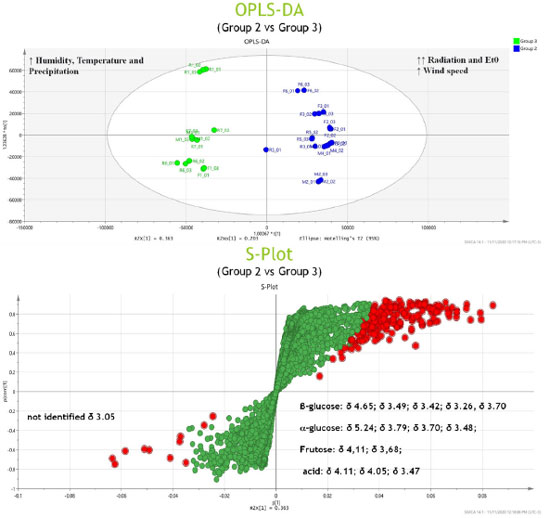
RESULTS AND DISCUSSION Analysis of metabolic profiles using NMR spectroscopy For our analysis, S. brasiliensis trees growing in three different microenvironments were selected. Thus, we initially identified the primary organic components in the aqueous extracts of bark samples collected from these different sites (forest, management, and reserve, Figure 1 (a)-(c)). A comprehensive analysis of the spectral data obtained using the 1H NMR and 2D J-resolved (J-res) spectroscopy techniques enabled the identification of compounds present in the extracts obtained from S. brasiliensis plant specimens. NMR feature assignments for the studied genera and families were substantiated by analyzing a previously generated database. The compounds in each species were comprehensively analyzed, and the results are presented in Table 1, and the representative 1H NMR, 1D J-res NMR, and 2D J-res NMR spectral profiles recorded for each plant extract were obtained. Four major chemical shift regions were identified in the NMR spectral profile recorded for the S. brasiliensis samples (Figure 1). Peaks appeared in the regions corresponding to the aliphatic (0.50-2.00 ppm), sugar and organic acid (3.00-6.00 ppm), amino acid (2.00-7.50 ppm), and aromatic (6.00-9.00 ppm) groups. The signals corresponding to the carbohydrate units, appearing in the range of 4.00-6.00 ppm, could be attributed to the anomeric protons in sugars. Additionally, the multiplicity and chemical shifts of these signals in the 1D and 2D J-res spectral profiles assisted in determining the nature of glycosides in the samples. The chemical shifts of the signals corresponding to protons in amino acids depended on the positions of the protons within the molecules and the nature of the side chains. Generally, signals corresponding to the aromatic protons in phenylalanine appear in the range of 6.0-8.5 ppm, and the chemical shift of the signal corresponding to the alpha proton in serine appears in the range of 3.8-4.0 ppm.
 Figure 1. 1H NMR spectral profiles recorded for aqueous bark extracts of S. brasiliensis grown in the forest (a), management (b), and reserve (c) sites. These 1H NMR spectral profiles were recorded using a 600 MHz spectrometer at a controlled temperature of 298 K
The NMR parameters adopted for identifying the compounds, such as 1H and 13C chemical shifts, coupling constants, and heteronuclear correlations, are listed in Table 1. Notably, acetic, lactic, malic, and quinic acids contain carboxylic acid groups. Hence, 1H and 13C chemical shifts recorded for these compounds are similar. These peaks generally appear in the region corresponding to organic acids (1.00-4.50 ppm for 1H and 160-180 ppm for the 13C NMR signals). The peaks corresponding to anomeric protons and carbons in the profiles recorded for fructose, α-glucose, β-glucose, and sucrose typically appear in the ranges of 4.5-6.0 ppm and 70-110 ppm, respectively. The differences in the chemical shifts can be attributed to the distinct stereochemistry of the monosaccharides and the glycosidic linkages in sucrose. Moreover, phenylalanine and serine bear different side chains. In phenylalanine, the peaks corresponding to aromatic protons typically appear in the range of 7.0-7.5 ppm, whereas the peaks corresponding to aromatic carbons generally appear between 120 and 140 ppm. The NMR signals corresponding to the alpha protons in serine appear in the range of 3.8-4.0 ppm, whereas the signals corresponding to the alpha carbons appear in the range of 50-60 ppm. The side chains of these amino acids, the phenyl ring in phenylalanine, and the hydroxyl group in serine contribute to variations in the chemical shifts. Sucrose, glucose and fructose are commonly found in barks. Carbohydrates are important back bones of life strategies of long-living trees, and barks are also the major storage compartments of carbon. In addition, during the heartwood formation, carbohydrates sustain the formation of phenolic extractives that ascribe for the natural durability of wood.35 Roots absorb phenylalanine and transported it to the aerial parts including wood, bark and leaves.36 In addition, the presence of amino acids as alanine and serine in barks may be associated to the origins of tannins and flavonoids in barks and heartwoods, such as for black-wattle.37 The presence of gallic, citric and quinic acids in the barks extracts was confirmed by the following study using LC-MS analysis developed by our research group.1 Chemometric analysis - part 1: analysis of the chemical data obtained from different microenvironments Initially, a comprehensive analysis of the entire dataset was conducted, and the results were presented as the average of the values recorded over three years during the collection of S. brasiliensis bark samples from different microenvironments. This approach ensured that the working matrix represented the mean intensity of the identified metabolite responses generated in the microenvironments over the three-year collection period. However, the specific effects of the collection timeframe were not considered during the analytical procedure. With this, we aimed to understand the differences in the metabolic expression levels among the microenvironments of the forest, management, and reserve sites. The PLS-DA-based supervised chemometric analysis method was used to overcome the challenges presented by visual inspections of the compositional variability in S. brasiliensis bark samples and to identify the compounds that imparted distinct characteristics to barks based on plant growth in the forest, management, and reserve sites. Figure 2 presents the score (a) and loading (b) data. The number of LVs (4) was chosen based on the captured variance in each LV (79.17% total), and the errors in terms of RMSEC and RMSECV decreased to 0.31 and 0.37, respectively (similarity index: 0.84, RMSEC/RMSECV).
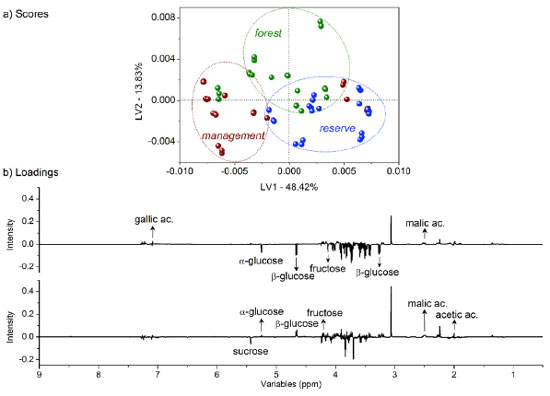 Figure 2. (a) LV1 × LV2 score coordinate system from the classification of barks based on tree growth characteristics: forest in green, management in red, and reserve in blue; (b) respective loadings with identified compounds based on their relevance according to the variable importance in the projection (VIP) analysis
Although some bark extracts were misclassified as outliers, the separation tendency of the bark extracts was observed to depend on the microenvironment. The LV1 axis was the primary factor for bark extract classification, accounting for 48.42% of the total variance. Bark extracts from the forest and management sites contained higher amounts of fructose and α- and β-glucose than those collected from the other site. By contrast, the bark extracts sourced from the reserve site contained higher concentrations of malic and gallic acids than the bark extracts collected from the other two sites. The LV2 axis played a significant role in identifying bark extracts with high contents (combined amounts) of the aforementioned compounds (exception: for sucrose, present in high concentrations in extracts from the management and reserve sites). Generally, primary metabolites are directly associated with physiological processes.38 The presence of high amounts of fructose and α- and β-glucose in the bark extracts collected from the forest and management sites could be potentially attributed to differences in environmental conditions, such as light availability, temperature, or nutrient levels.39 These factors can influence plant metabolism and growth, leading to variations in the amounts of primary metabolites such as sugars.40 Moreover, the pathway of primary metabolism may potentially change in response to adverse abiotic factors, as a series of adaptive mechanisms help plants evade and withstand these stresses. These mechanisms can also protect plants from stressful conditions, and related adjustments involve alterations in sugar, sugar alcohol, and amino acid levels depending on the species.16 Some specialized metabolites are synthesized from the limited products of primary metabolism, indicating a correlation between the two types of metabolic processes. Some resources are used for growth, while the rest are used for environmental adaptation.8,41 The presence of high concentrations of malic and gallic acids in the bark extracts obtained from the reserve region could be an indication of the adaptive responses of trees to specific environmental conditions or stresses. Malic acid is associated with the production of energy in plants and can help plants cope with environmental stresses such as drought or nutrient deficiency.42 Gallic acid, a phenolic compound with antioxidant properties, can protect plants from oxidative stress attributable to various biotic and abiotic factors.43 The main classes of specialized metabolites are generated during the progress of different primary metabolite pathways, such as glycolysis, Krebs cycle, pentose phosphate pathways, and shikimate pathways.41 Chemometric analysis - part 2: analysis of dendrometric and environmental data recorded in the different microenvironments Dendrometry, the study of tree dimensions, is critical for understanding forest dynamics and assessing the effects of various microenvironments on the rate of tree growth and productivity.44 The analysis of dendrometric data obtained from different microenvironments provides insights into the influence of factors, such as climate, soil type, topography, and species composition, on tree growth patterns and forest structure.45 Consequently, by analyzing these data, researchers can study the adaptive responses of trees to their microenvironments, identify factors that promote optimal growth, and develop management strategies that promote the development of sustainable forest ecosystems.46 Thus, the exploration of dendrometric data across diverse microenvironments is essential to develop a comprehensive understanding of forest ecology. The data can help develop effective forest management practices.47 In our analysis, gum formation was observed in the barks during sample collection. These substances are produced in several plants that grow under semi-arid conditions. These plants produce gummy exudates in large quantities to defend against dehydration.48 The composition of these gums is highly heterogeneous, and these are predominantly characterized by a substantial carbohydrate content. The carbohydrate content varies significantly and lies in the range of 43.51-98.46%. The compounds are polysaccharides in nature.49 This is heavily dependent on weather conditions.48 Following the multivariate classification analysis of microenvironments, regression modeling analysis methods were used to determine significant correlations between all compounds identified by analyzing the 1H NMR spectral profiles and previously studied dendrometric, environmental, and chemical variables. Data on several environmental factors such as mean temperature, relative humidity, incident solar radiation content, amount of precipitation, wind speed, and reference evapotranspiration were collected. Tree height, ground-level diameter, and treetop diameter constituted the dendrometric data. PLS-based multivariate regression analyses were conducted to correlate each of these variables (categorical variables; Y matrix) with the complete 1H NMR spectral profile (δ 0.5-9.0 ppm). The correlations established between the dendrometric and environmental data revealed consistent parameter correlations across the different microenvironments. Figure 3 presents data on the statistical parameters associated with modeling. Eight LVs for the environmental mean temperature (a), wind speed (b), reference evapotranspiration (c), tree height (d), diameter at ground level (e), diameter of the treetop (f), and their respective ideal (green) and actual (red) regression curves were established.
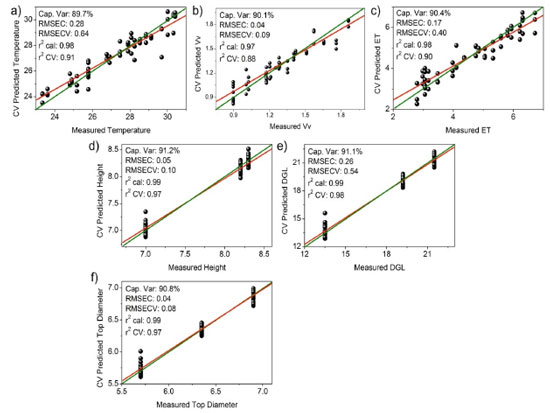 Figure 3. Ideal (green) and real (red) regression curves generated for multivariate modeling (95% confidence limit) based on the correlation of the 1H NMR dataset (δ 0.5-9.0 ppm) with the variables: (a) environmental mean temperature (ºC), (b) wind speed (m s-1), (c) reference evapotranspiration (ET), (d) tree height (m), (e) diameter at ground level (DGL) (cm), and (f) diameter of the top tree (m). Statistical parameters represented by the total captured variance (%), errors determined based on the RMSEC and RMSECV, and correlation coefficients (r2) obtained post calibration and cross-validation analysis
The obtained results can be explained by the fact that the tree height (d), diameter at ground level (e), and tree top diameter (f) exhibited the strongest correlations with all the compounds reflected by the 1H NMR spectral profiles. This can be potentially attributed to the fact that these dendrometric parameters are directly related to the growth and overall health of trees.50 Therefore, these parameters reflect the ability of trees to produce and accumulate various chemical compounds under the influence of various environmental factors. However, variables such as biomass, relative humidity of air, incident solar radiation content, and precipitation did not correlate with the composition of the extracts collected from the three different sites mentioned earlier. This lack of correlation could be attributed to the complex and indirect influence of these variables (except for biomass) on the chemical composition of the tree bark samples.39 We did not expect the biomass to correlate with the chemical profile of the trees, as biomass was quantified based on an allometric calculation method using a single value (the diameter of the trunk at ground level). Therefore, the biomass was expected to be directly related to the diameter in the correlation. This lack of correlation could be potentially attributed to the use of an exponential equation. The use of such an equation may have masked the interdependence between diameter and biomass. These variables can potentially influence multiple aspects of tree growth and physiology, and understanding their effects on the chemical composition of trees based on data may be difficult.51 Additionally, interactions between these variables and other environmental factors could further complicate the relationship between these variables and the chemical composition of the extracts.52 The errors represented by RMSEC and RMSECV, the relatively high values of calibration and cross-validation coefficients (r2), and the proximity between the ideal (green) and real (red) regression curves indicate well-adjusted models. Furthermore, the models for tree height (d), diameter at ground level (e), and diameter of the top tree (f) exhibited the best correlation with all compounds reflected by the 1H NMR spectral profiles.25 The other variables (biomass, relative air humidity, incident solar radiation content, and precipitation) did not correlate satisfactorily with the composition of the extracts obtained from the three different environments (data not shown). Chemometric analysis - part 3: analysis of correlation between collection periods and environmental factors As stated, plant material from S. brasiliensis was collected between 2015 and 2018. Seven collection cycles were conducted across four seasons in three distinct microenvironments. The CCA enabled the correlation of bark collection periods in the three microenvironments and environmental variables with the metabolites present in the samples. Results obtained from the CCA (Figure 4) demonstrated that each environmental factor exhibited distinct effects on the metabolites extracted from the barks of S. brasiliensis. The samples were categorized into four groups, and the formation of these groups was based on the similarity between the environment corresponding to the collection period and the metabolites present.
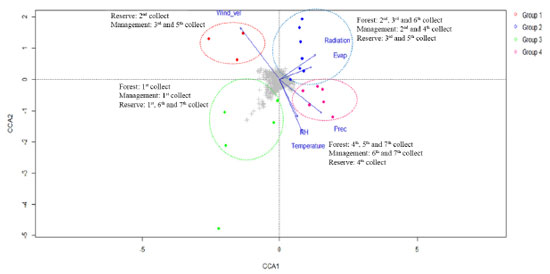 Figure 4. CCA results on the metabolic fingerprint obtained by analyzing NMR profiles of S. brasiliensis bark extracts and the data corresponding to the correlation between environmental factors (and collection periods) and the cluster of metabolic similarity
The members of Group 1, positioned in the negative region of CCA 1 (Figure 4) and positive region of CCA 2 (Figure 4), were primarily affected by the wind speed and exposed to intermediate levels of radiation and evapotranspiration. This group consisted of samples collected during the 2nd collection cycle from the reserve, as well as the 3rd and 5th collection cycles from the management site. The samples were primarily collected in June. By contrast, the members of Group 4 were concentrated in the positive region of CCA 1 and the negative region of CCA 2 and were exposed to intermediate and high levels of precipitation, temperature, and humidity. This group contained samples collected during the 4th, 5th, and 7th collection cycles from the forest; the 6th and 7th collection cycles from the management site; and the 4th collection cycle from the reserve. Notably, this group primarily consisted of samples collected during the time period ranging from February 2017 to February 2018 when high precipitation rates were recorded. Group 2, exposed to the highest levels of radiation and evapotranspiration and intermediate levels of wind speed, was positive in terms of both CCA 1 and CCA 2. This group consisted of samples collected during the 2nd, 3rd, and 6th collection cycles from the forest; the 2nd and 4th collection cycles from the management site; and the 3rd and 5th collection cycles from the reserve. The samples were primarily collected between June and October. The members of Group 3 were concentrated in the negative regions of CCA 1 and CCA 2 and exposed to low levels of radiation and temperature and intermediate levels of precipitation, humidity, and temperature. This group consisted of samples from the 1st collection from the forest; the 1st collection from the management site; and the 1st, 6th, and 7th collections from the reserve. The majority of the samples belonging to this group were collected in February 2016. Results from the CCA revealed that all the groups comprised samples from the three microenvironments (reserve, management, and forest), and the collection period corresponding to each sample group differed from the others. The responses of different microenvironments were recorded over the same time period, and the responses generated were observed to be distinctly different from each other. For example, collection 5 was included in three groups: the forest in Group 1, the reserve in Group 2, and the management site in Group 4. The results revealed that each environmental and field situation responded differently to different environmental factors, and this affected the chemical profile of trees. The results indicated a correlation between genetic and environmental variability. The PCA (Figure 5) was performed in conjunction with the OPLS-DA and S-plot analysis (Figures 6(a)-(b) and 7(c)-(d)). Chemical identification based on loading plots obtained post PCA allowed a tentative classification of the substance classes, as high dimensional 1H NMR data were recorded. Consequently, we opted to use the OPLS-DA method and S-plot models to accurately identify the chemical markers. The data were subjected to PCA using the pareto scale. The PCA-X scores and loading graph represented 58.73% of the total variance (R2X[1] = 0.381 and R2X[2] = 0.206). The 63 samples were categorized into four groups based on the collection cycle/area of collection. Group 1, located in the positive region of PC 1 and the negative region of PC 2, consisted of samples from the 2nd collection from the reserve and the 3rd and 5th collections from the management site. Group 2, located in the positive regions of PC 1 and PC 2, comprised of samples from the 2nd, 3rd, and 6th collections from the forest; the 2nd and 4th collections from the management site; and the 3rd and 5th collections from the reserve. By contrast, Group 3, located in the negative regions of PC 1 and PC 2, comprised of samples from the 1st collection from the forest; the 1st collection from the management site; and the 1st, 6th, and 7th collections from the reserve. Finally, Group 4, positioned in the negative region of PC 1 and the positive region of PC 2, included samples from the 4th, 5th, and 7th collections from the forest; the 6th and 7th collections from the management site; and the 4th collection from the reserve.
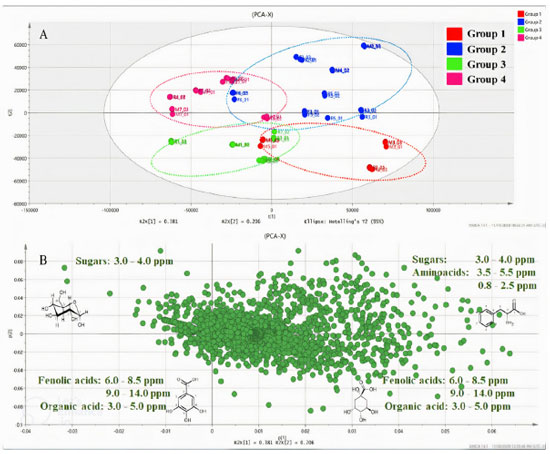 Figure 5. Graph presenting the PCA score (A) and loading (B) data for S. brasiliensis bark samples obtained from three microenvironments for the seven samples analyzed using the NMR spectroscopy technique
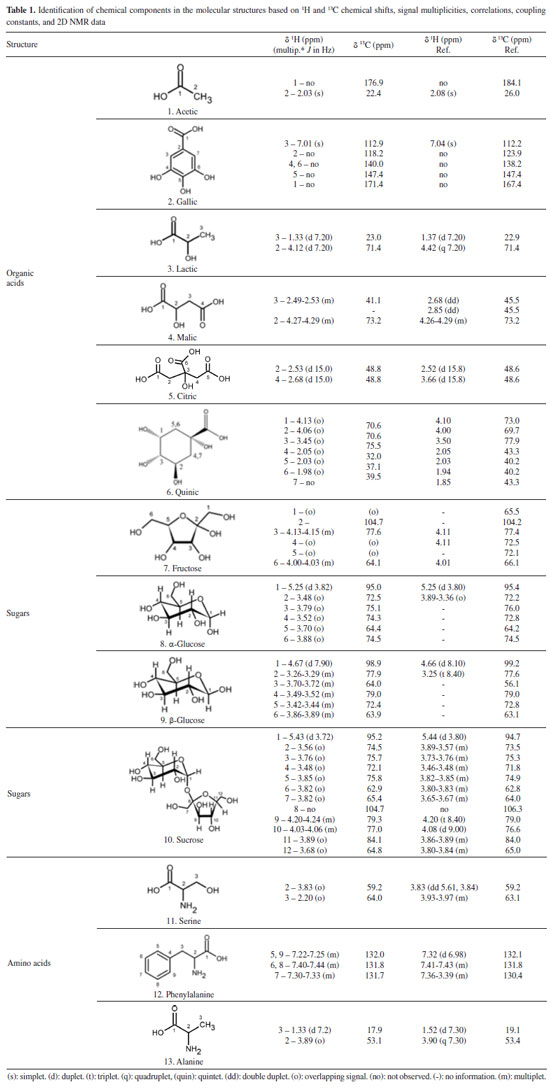
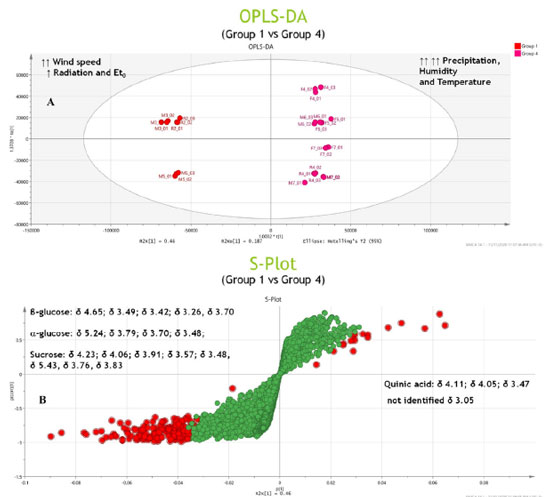
Potential markers The PCA results (Figure 5) revealed the distinct formation of four clusters based on the collection period and environmental factors. Each group was characterized by specific primary metabolites. The OPLS-DA was performed using VIP scores to identify possible discriminant metabolites that accounted for the differences between the groups. VIP values greater than three and p values less than 0.05 were considered significant. Figure 6 (A and B) presents the OPLS-DA results obtained by comparing opposing groups: Group 1 vs. Group 4 (R2 = 0.992, Q2 = 0.988) and Group 2 vs. Group 3 (R2 = 0.956, Q2 = 0.946). S-plots, Figure 7 (C and D) were generated from the OPLS-DA data, and several potential biomarkers were selected (Table 1).
Group 1, consisting of samples from the 2nd collection from the reserve and the 3rd and 5th collections from the management site, was significantly influenced by the wind speed, intermediate levels of radiation and evapotranspiration, and low levels of precipitation. Sugars such as glucose, α-glucose, β-glucose, fructose, and sucrose accumulated in the samples in this group. The results obtained during the first part of the chemometric analysis process revealed that samples from the reserve and management microenvironments exhibited higher concentrations of sugars than those from the forest microenvironment. The results agreed well with the results obtained during the second part of the analytical process, where it was observed that Group 1 consisted of clusters that contained sugars (particularly sucrose) as discriminating metabolites. The nature of the sugars depended on the collection period. As sessile organisms, plants cannot evade the extreme conditions of the environment in which they are rooted. Consequently, they develop highly complex mechanisms that are based on the features of physiology and metabolism to adapt to the conditions they are exposed to.8,24 Several factors, such as water deficit, irradiation amount and intensity, and temperature conditions, simultaneously affect plants. Thus, identifying the factor primarily responsible for generating a given physiological response is often difficult.10,53 Plants are susceptible to mechanical damage caused by various factors (such as wind). This can influence the expression of secondary metabolism.54,55 The primary biosynthetic pathways associated with the generation of these metabolites are derived from primary metabolism pathways. In other words, changes in the properties of one type of metabolism influence the properties of another.8,9 Next, water stress has been reported to significantly affect the nature of metabolites produced by plants. Such water stress can result in an increase in the concentration of specific amino acids, sugars, and their derivatives.56 Under drought conditions, sugars accumulate first, followed by amino acids. The accumulation of sugars, such as glucose, fructose, galactose, and sucrose, results in inhibited activity of hexokinase. This helps in regulating the osmotic potential of the cells, thereby safeguarding the cells from osmotic stress during drought conditions. In certain cases, plants do not acclimatize and begin to synthesize various compounds, including amino acids (such as tyrosine and proline) and specialized metabolites.57 Thus, various bioactive compounds are synthesized by plants under these conditions to cope with water deficit.58 During this period, plants may present an increase in phosphoenolpyruvate carboxylase activity. Such increase in activity indicates the formation of specialized metabolites from sucrose,58 which is the primary sugar transported in the largest plant species and is known to accumulate under stress.57 Group 4, which was subjected to high levels of precipitation and was significantly influenced by temperature and humidity, consisted of samples from the 4th, 5th, 7th collections from the forest; 6th and 7th collections from the management site; and the 4th collection from the reserve site. Organic acids (such as quinic and acetic acid) and an unidentified compound (δ 3.05 ppm) were present in high concentrations in this group. Generally, the metabolism of organic acids is associated with environmental stress. This metabolic pathway serves as an intermediary pathway in carbon metabolism and plays pivotal roles in several other processes that help some plants withstand various stressful conditions (such as nutrient deficiency, herbivory, and pathogen attack). These mechanisms also help plants achieve metal tolerance.42 Generally, heavy rainfall and an increase in the water content of soil above field capacity cause problems, as oxygen availability to the roots is reduced under such conditions.10 Depending on the biomass and microbiota in the soil, such reduction in O2 availability can occur within 24 h.10 Under these conditions, adenosine triphosphate must be produced during fermentation. This can result in cytosolic acidification and the accumulation of toxic products. Moreover, the accumulation of amino acids and low levels of sugars and various organic acids can also be achieved under these conditions.57 In addition to precipitation, temperature significantly influences plant growth and development rates as it affects several highly sensitive biochemical reactions.59 The stability of proteins and nucleic acids, structure of the cytoskeleton, and efficiency of enzymatic reactions are significantly affected by heat stress under conditions of high and low temperatures, resulting in a metabolic imbalance.10 High temperature and humidity often increase the enzyme content and induce enzymes that sequester reactive oxygen species, causing various physiological, biochemical, and molecular changes in plant metabolism.8,60 To counterbalance the biochemical and physiological alterations induced by stress, various key mechanisms of tolerance have been employed, including the utilization of ion transporters, proteins, osmoprotectants, antioxidants, and other factors associated with signaling and transcriptional control.59 Moreover, a profound change in the pool of metabolites in the affected plants is observed with changes in various environmental factors (such as temperature, humidity, and water availability).53,54,57,61 For Group 2, S-plots, Figure 7 (C and D) were generated from the OPLS-DA data, and some possible biomarkers were selected (Table 1). High concentrations of sugars (such as α-glucose and β-glucose) and the amino acid serine were present in this group consisting of samples from the 2nd, 3rd, and 6th collections from the forest; 2nd and 4th collections from the management; and 3rd and 5th collections from the reserve sites exposed to conditions of intense solar radiation and high rates of evapotranspiration. Notably, solar radiation is an essential and influential environmental factor that affects plant growth and development. Plant survival depends on the ability of plants to accumulate biomass and fix carbon through photosynthesis. Plants develop evolved mechanisms that protect them against excessive ultraviolet radiation (UV) and detrimental light conditions.8 Elevated radiation intensities trigger the production of reactive oxygen species, and this, in turn, results in the stimulation of protective responses. These responses promote the biosynthesis of specialized metabolites capable of absorbing UV-B radiation. This eventually results in alterations in the activities of antioxidant enzymes.58 High levels of radiation have been observed to correlate positively with the production of phenolic compounds,54 and most phenolic compounds in plants are derived from amino acids, such as phenylalanine. Conversely, Group 3, exposed to intermediate temperature and humidity levels, consists of samples from the 1st collection from the forest; the 1st collection from the management; and the 1st, 6th, and 7th collections from the reserve sites. Potential organic acid markers (such as gallic and quinic acid) and an unidentified compound (δ 3.05 ppm), which were absent in Group 2, were identified in this group. The high radiation intensities and low precipitation levels were associated with an increase in the concentrations of sugars and amino acids. Furthermore, each microenvironment generated a distinct response to the various environmental factors under investigation. Quantification of metabolites in the barks of S. brasiliensis using 1H qNMR spectroscopy Figure 8 depicts bar graphs presenting data on the compounds quantified using the 1H qNMR spectroscopy technique. The bar graphs reflect data related to the areas of collection and the collection periods. The results obtained using this quantitative analysis method supported and complemented the results obtained post chemometric analyzes. ANOVA tests were conducted for each area over the seven collection periods. The results revealed that the samples were statistically different with respect to the concentration of each compound.
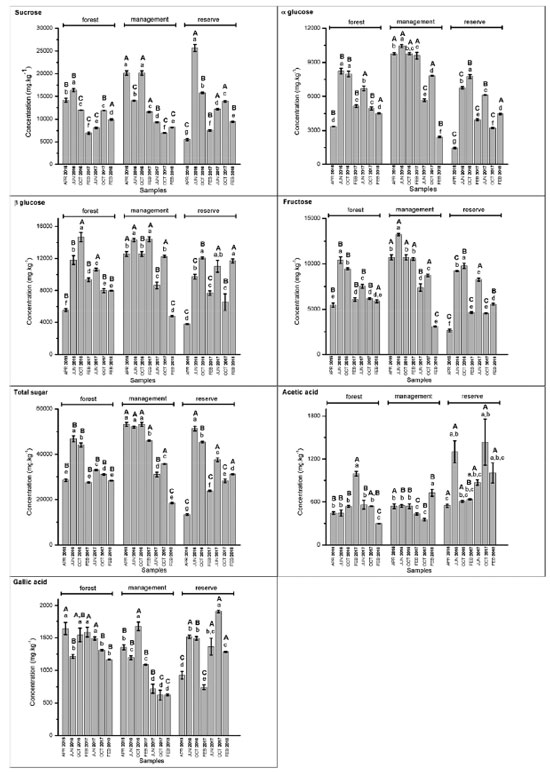 Figure 8. Concentrations (mg kg-1) of sucrose, α-glucose, β-glucose, fructose, total sugars, acetic acid, and gallic acid determined based on the period and area of collection of S. brasiliensis barks using 1H qNMR spectroscopy. Capital letters refer to intragroup ANOVA results, and lowercase letters refer to intergroup ANOVA results for each compound
The results obtained from this quantification analysis enabled us to correlate the groups formed based on the CCA results (Figure 4) with those formed based on the PCA results (Figure 5). Sucrose was detected in high concentrations in Group 1 (Figure 8). This was observed by analyzing the samples collected in June 2016 from the reserve and those collected in October 2016 from the management site. The lowest concentrations of sugars, such as sucrose, and the highest concentrations of acetic acid (Figure 8) were recorded for Group 4. The results were obtained by analyzing the collections made in February 2017 (forest and reserve), February 2018 (forest and management), June 2017 (forest), and October 2017 (management). The results agreed well with the results presented in papers that reported increased sucrose levels in plants under drought stress conditions. Notably, sucrose serves as a compatible solute and osmoprotectant.62 Low sugar concentrations and high acetic acid levels were recorded for Group 4, and the results agreed well with the results reported in the literature on the role of acetic acid in the activation of pathways associated with drought stress response.63 High concentrations of sugars (such as glucose and glucose) were recorded for Group 2, which comprised of samples collected in June 2016 (forest and management), October 2016 (forest and reserve), February 2017 (management), June 2017 (reserve), and October 2017 (forest) (Figure 7). Group 3 samples contained gallic acid as the discriminating metabolite (Figure 7). This group consisted of samples from the 1st collection (April 2016) from the three areas and the 6th (October 2017) and 7th (February 2018) collections from the reserve site. Notably, the above sugars have been reported to accumulate in response to abiotic stress and promote osmotic adjustment and stress tolerance in plants.64 The presence of gallic acid as a discriminating metabolite in Group 3 aligns with the results reported in papers highlighting the role of gallic acid as an antioxidant. Moreover, gallic acid has been reported to help plants cope with oxidative stress attributable to environmental changes.65 Above-average concentrations of total sugars (sucrose, fructose, glucose, and glucose) were detected in the samples collected from the management (April, June and October 2016, and February 2017), reserve (June and October 2016, and June 2017), and forest (June and October 2016) areas. The samples collected from each area elicits distinct responses to various environmental stimuli, regardless of whether it is during the rainy or dry season. These responses manifest as variations in sugar concentrations, with different areas presenting high sugar concentrations at different times of the year. A significant increase in the total sugar concentration was recorded during the drought periods in the samples collected from the reserve and forest areas in June and October 2016, and June 2017. A considerable variability was noted between areas during the same collection period. The observed variability in total sugar concentrations (in response to drought conditions) across areas and collection periods reflects the plasticity of plant metabolic responses to fluctuating environmental conditions.66 Plasticity is crucial for plant survival and plant adaptation to extreme environmental conditions. The survival of plants in semi-arid Brazilian climate and desert regions is influenced by plasticity.67-69 In summary, the results reported herein agree well with those reported in existing literature reports concerning plant metabolic responses to environmental stress, particularly under extreme climatic conditions. Further research should, however, be conducted to help elucidate the specific metabolic pathways and regulatory mechanisms associated with the generation of these responses. The obtained results can potentially help us better understand plant adaptation and resilience strategies.
CONCLUSION This paper reports findings indicating a correlation between the metabolic profiles of S. brasiliensis samples collected from three microenvironments. The results were influenced by the sampling period and prevailing environmental conditions. The analytical approach followed, coupled with the use of a suitable multivariate analysis method, facilitated the generation of robust and reliable statistical results. The approach contributed to the comprehensive understanding of the influence of certain specialized metabolites, particularly primary metabolites in specimens, depending on different microenvironmental conditions. Consequently, changes in the abiotic environmental factors describing the selected locations were observed to affect the nature and content of the metabolites. The methodology followed can be potentially used to investigate various species of interest, particularly in the context of reforesting degraded regions within the Caatinga biome using highly adaptable species. Additionally, it can be used to explore the effects of diverse cultivation areas and environmental changes on plant development. Moreover, the reported approach can be further developed to examine the quality control and biological properties of other economically important crops and medicinal plants. We can infer that the results reported herein provide valuable insights into the chemical ecology of important plant species.
SUPPLEMENTARY MATERIAL Figure 1S illustrates the changes in the daily average wind speed (m s-1). Figure 2S illustrates the behavior of the daily total reference evapotranspiration (ETo), determined based on the FAO Penman-Monteith equation and the daily global solar radiation (Rg). Figure 3S illustrates the behavior of the maximum (Tmax) and minimum (Tmin) air temperature and total daily precipitation (mm). Figure 4S illustrate the behavior of the average relative humidity (RH avg), observed at the Caatinga Meteorological Station from 2016 to 2018 (Petrolina, PE). All of them are available at http://quimicanova.sbq.org.br, in the form of a PDF file, with access free.
ACKNOWLEDGMENTS The authors gratefully acknowledge the financial support from Embrapa (SEG 03.14.01.012.00.00), National Council for Scientific and Technological Development (CNPq, Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES, National Institute of Science and Technology - INCT BioNat (grant # 465637/2014-0), Brazil. The authors thank CNPq for financial support and scholarships (303791/2016-0 and DCR-0024-01686.01.00/15).
REFERENCES 1. Luz, L. R.; Porto, D. D.; Castro, C. B.; Silva, M. F. S.; Alves Filho, E. G.; Canuto, K. M.; de Brito, E. S.; Becker, H.; do Ó Pessoa, C.; Zocolo, G. J.; J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018, 1099, 97. [Crossref] 2. Fernandes, F. H. A.; de Batista, R. S. A.; de Medeiros, F. D.; Santos, F. S.; Medeiros, A. C. D.; Rev. Bras. Farmacogn. 2015, 25, 208. [Crossref] 3. Santos, C. C. S.; Masullo, M.; Cerulli, A.; Mari, A.; Estevam, C. D. S.; Pizza, C.; Piacente, S.; Phytochemistry 2017, 140, 45. [Crossref] 4. Saraiva, A. M.; Saraiva, C. L.; Cordeiro, R. P.; Soares, R. R.; Xavier, H. S.; Caetano, N.; Rev. Bras. Plant. Med. 2013, 15, 199. [Crossref] 5. Sette-de-Souza, P. H.; Medeiros, F. D.; Santana, C. P.; Araújo, R. M.; Cartaxo-Furtado, N. A. O.; Macêdo, R. O.; Medeiros, A. C. D.; J. Therm. Anal. Calorim. 2018, 131, 829. [Crossref] 6. Parente, E.; Plantas de Valor Econômico no Ceará; O Ceará: Fortaleza, 1966. 7. Andrade-Lima, D.; Plantas da Caatinga, Rio de Janeiro: Academia Brasileira de Ciências, 1989. 8. Alnsour, M.; Ludwig-Müller, J.; Journal of Endocytobiosis and Cell Research 2015, 26, 90. [Link] accessed in August 2023 9. Carril, E. P. U.; Ávalos García, A.; Reduca 2009, 2, 119. [Link] accessed in August 2023 10. Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A.; Int. J. Mol. Sci. 2013, 14, 4885. [Crossref] 11. Chaves, M. M.; Flexas, J.; Pinheiro, C.; Ann. Bot. 2009, 103, 551. [Crossref] 12. Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R.; Physiol. Plant. 2008, 132, 199. [Crossref] 13. Ramakrishna, A.; Ravishankar, G. A.; Plant Signaling Behav. 2011, 6, 1720. [Crossref] 14. Sampaio, B. L.; Edrada-Ebel, R.; da Costa, F. B.; Sci. Rep. 2016, 6, 29265. [Crossref] 15. Sampaio, B. L.; da Costa, F. B.; Rev. Bras. Farmacogn. 2018, 28, 135. [Crossref] 16. Canuto, G. A. B.; da Costa, J. L.; da Cruz, P. L. R.; de Souza, A. R. L.; Faccio, A. T.; Klassenc, A.; Rodrigues, K. T.; Tavares, M. F. M.; Quim. Nova 2018, 41, 75. [Crossref] 17. Krishnan, P.; Kruger, N. J.; Ratcliffe, R. G.; J. Exp. Bot. 2005, 56, 255. [Crossref] 18. Freitas, J. V. B.; Alves Filho, E. G.; Silva, L. M. A.; Zocolo, G. J.; de Brito, E. S.; Gramosa, N. V.; Talanta 2018, 180, 329. [Crossref] 19. Oliveira, V. R.; Ribeiro, A.; Pareyn, F. G. C.; Drumond, M. A.; Porto, D. D.; Kiill, L. H. P. F. F.; A. C. Rev. Cern. 2023, 29, e-103165. [Crossref] 20. Lima, P. C. F.; de Souza, S. M.; Drumond, M. A.; Edição dos Anais do 5º Congresso Nacional sobre Essencias Nativas, Campos do Jordão, Brasil, 1982. [Link] accessed in August 2023 21. https://www.cbd.int/abs/about/, accessed in August 2023. 22. de Moura, M. S. B.; Dados Climáticos Estação Meteorológica do Campo Experimental da Caatinga, 2005, 1ª ed.; Embrapa: Petrolina, 2007. [Link] accessed in August 2023 23. Silva, L. M. A.; Alves Filho, E. G.; Choze, R.; Lião, L. M.; Honda, N. K.; Alcantara, G. B.; J. Braz. Chem. Soc. 2012, 23, 273. [Crossref] 24. Ahuja, I.; de Vos, R. C. H.; Bones, A. M.; Hall, R. D.; Trends Plant Sci. 2010, 15, 12. [Crossref] 25. Alves Filho, E. G.; de Brito, E. S.; Rodrigues, S. In Advances in Cold Plasma Applications for Food Safety and Preservation; Aguirre, D. B. , ed.; Academic Press: Cambridge, 2020, ch. 8. 26. Galili, S.; Amir, R.; Galili, G.; Adv. Plant Biochem. Mol. Biol. 2008, 1, 49. [Crossref] 27. Origin, version 9.4; OriginLab Corporation; Northampton, MA, USA, 2017. 28. Matlab®, version R2013a; The MathWorks Inc.; Natick, MA, USA, 2013. 29. PLS Toolbox, version 8.6.1; Eigenvector Research Inc.; Wenatchee, USA, 2018. 30. Garcia, E.; Klaas, I.; Amigo, J. M.; Bro, R.; Enevoldsen, C.; J. Dairy Sci. 2014, 97, 12. [Crossref] 31. Guedes, J. A. C.; Alves Filho, E. G.; Rodrigues, T. H. S.; Silva, M. F. S.; Souza, F. V. D.; Silva, L. M. A.; Alves, R. E.; Canuto, K. M.; de Brito, E. S.; Pessoa, C. O.; Nascimento, R. F.; Zocolo, G. J.; Ind. Crops Prod. 2018, 124, 466. [Crossref] 32. R, version 4.0.3; R Foundation for Statistical Computing; Austria, 2020. 33. SIMCA, version 14.1; Umetrics; Sweden, 2017. 34. Sucupira, N. R.; Alves Filho, E. G.; Silva, L. M. A.; de Brito, E. S.; Wurlitzer, N. J.; Sousa, P. H. M.; Food Chem. 2017, 216, 217. [Crossref] 35. Magel, E.; Einig, W.; Hampp, R.; Dev. Crop Sci. 2000, 26, 317. [Crossref] 36. Jiao, Y.; Chen, Y.; Ma, C.; Qin, J.; Nguyen, T. H. N.; Liu, D.; Gan, H.; Ding, S.; Luo, Z. B.; Tree Physiol. 2018, 38, 66. [Crossref] 37. Masendra; Ashitani, T.; Takahashi, K.; Susanto, M.; Lukmandaru, G.; Journal of the Korean Wood Science and Technology 2019, 47, 80. [Crossref] 38. Deborde, C.; Moing, A.; Roch, L.; Jacob, D.; Rolin, D.; Giraudeau, P.; Prog. Nucl. Magn. Reson. Spectrosc. 2017, 102-103, 61. [Crossref] 39. Etherington, J. R.; Larcher, W.; J. Ecol. 1996, 84, 630. [Crossref] 40. Caretto, S.; Linsalata, V.; Colella, G.; Mita, G.; Lattanzio, V.; Int. J. Mol. Sci. 2015, 16, 26378. [Crossref] 41. Aharoni, A.; Galili, G.; Curr. Opin. Biotechnol. 2011, 22, 239. [Crossref] 42. López-Bucio, J.; Nieto-Jacobo, M. F.; Ramírez-Rodríguez, V.; Herrera-Estrella, L.; Plant Sci. 2000, 160, 1. [Crossref] 43. Badhani, B.; Sharma, N.; Kakkar, R.; RSC Adv. 2015, 5, 27540. [Crossref] 44. Kershaw, J. A. J.; Ducey, M. J.; Beers, T. W.; Husch, B.; Forest Measurements, 5th ed.; McGraw-Hill: New York, 2009. 45. Chave, J.; Andalo, C.; Brown, S.; Cairns, M. A.; Chambers, J. Q.; Eamus, D.; Fölster, H.; Fromard, F.; Higuchi, N.; Kira, T.; Lescure, J. P.; Nelson, B. W.; Ogawa, H.; Puig, H.; Riéra, B.; Yamakura, T.; Oecologia 2005, 145, 87. [Crossref] 46. Pretzsch, H.; Forrester, D. I.; Rötzer, T.; Ecol. Modell. 2015, 313, 276. [Crossref] 47. Bontemps, J. D.; Bouriaud, O.; Forestry 2014, 87, 109. [Crossref] 48. Robbers, J. E.; Speedie, M. K.; Tyler, V. E.; Pharmacognosy and Pharmacobiotechnology, 2nd ed.; Williams & Wilkins: New Delhi, 1997. 49. Hamdani, A. M.; Wani, I. A.; Bhat, N. A.; Int. J. Biol. Macromol. 2019, 135, 46. [Crossref] 50. Pretzsch, H.; Forest Dynamics, Growth and Yield: From Measurement to Model, 1st ed.; Springer: Berlin, 2010. 51. De Schepper, V.; Steppe, K.; Ann. Bot. 2011, 108, 1147. [Crossref] 52. Way, D. A.; Oren, R.; Tree Physiol. 2010, 30, 669. [Crossref] 53. Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R.; Plant Physiol. 2004, 134, 1683. [Crossref] 54. Gobbo-Neto, L.; Lopes, N. P.; Quim. Nova 2007, 30, 374. [Crossref] 55. Pereira, A.; Front. Plant Sci. 2016, 7, 1123. [Crossref] 56. Babita, M.; Maheswari, M.; Rao, L. M.; Shanker, A. K.; Rao, D. G.; Environ. Exp. Bot. 2010, 69, 243. [Crossref] 57. Obata, T.; Fernie, A. R.; Cell. Mol. Life Sci. 2012, 69, 3225. [Crossref] 58. Borges, C. V.; Minatel, I. O.; Gomez-Gomez, H. A.; Lima, G. P. P. In Medicinal Plants: Influence of Environmental Factors on the Content of Secondary Metabolites; Ghorbanpour, M.; Varma, A. , eds.; 2017, p. 259. [Crossref] 59. Hasanuzzaman, M.; Nahar, K.; Alam, M. M.; Roychowdhury, R.; Fujita, M.; Int. J. Mol. Sci. 2013, 14, 9643. [Crossref] 60. Padilla-González, G. F.; Frey, M.; Gómez-Zeledón, J.; da Costa, F. B.; Spring, O.; Sci. Rep. 2019, 9, 13178. [Crossref] 61. Borges, L. R.; Santos, D. C.; Cavalcanti, V. A. L. B.; Gomes, E. W. F.; Falcão, H. M.; da Silva, D. M. P.; Acta Hortic. 2013, 995, 359. [Crossref] 62. Yadav, B.; Jogawat, A.; Rahman, M. S.; Narayan, O. P.; Gene Rep. 2021, 23, 101040. [Crossref] 63. Utsumi, Y.; Utsumi, C.; Tanaka, M.; Van Ha, C.; Takahashi, S.; Matsui, A.; Matsunaga, T. M.; Matsunaga, S.; Kanno, Y.; Seo, M.; Okamoto, Y.; Moriya, E.; Seki, M.; Front. Plant Sci. 2019, 10, 1. [Crossref] 64. Martínez-Noël, G. M. A.; Tognetti, J. A. In Plant Metabolites and Regulation Under Environmental Stress; Ahmad, P.; Ahanger, M. A.; Singh, V. P.; Tripathi, D. K.; Alam, P.; Alyemeni, M. N. , eds.; Academic Press: Cambridge, 2018, p. 397. [Crossref] 65. Ozfidan-Konakci, C.; Yildiztugay, E.; Kucukoduk, M.; Environ. Sci. Pollut. Res. 2015, 22, 1487. [Crossref] 66. Shaar-Moshe, L.; Hayouka, R.; Roessner, U.; Peleg, Z.; Plant Direct 2019, 3, 1. [Crossref] 67. Silva, J. L. S. E.; Cruz-Neto, O.; Peres, C. A.; Tabarelli, M.; Lopes, A. V.; PLoS One 2019, 14, 1. [Crossref] 68. Souza, M. L.; Duarte, A. A.; Lovato, M. B.; Fagundes, M.; Valladares, F.; Lemos-Filho, J. P.; PLoS One 2018, 13, 1. [Crossref] 69. Lázaro-Nogal, A.; Matesanz, S.; Godoy, A.; Pérez-Trautman, F.; Gianoli, E.; Valladares, F.; J. Ecol. 2015, 103, 338. [Crossref] |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access