Artigo
| Thermoresponsive cellulose nanocrystals protecting macrophage cells from oxidative stress |
|
José A. PinheiroI; Nívia do N. MarquesI; Maylla M. C. L. SilvaII; Hugo A. O. RochaII; Rosangela de C. BalabanI,*
I. Instituto de Química, Universidade Federal do Rio Grande do Norte, 59078-970 Natal - RN, Brasil Recebido em 27/03/2023 *e-mail: rosangela.balaban@ufrn.br Cellulose nanocrystals (CNCs) were grafted with poly(N-isopropylacrylamide) (PNIPAM) chains, by means of the grafting from route, using the TEMED/KPS redox pair as free-radical initiators. The confirmation of grafting was revealed by 13C NMR spectra. In a test with murine macrophages (RAW 264.7 cell line), CNC-g-PNIPAM copolymers did not affect cell viability nor RAW cell oxide nitric production. Raw cells exposed to oxidative stress induced by H2O2 decreased the cell viability (~ 60%) and increased lipid peroxidation, as revealed by malondialdehyde (MDA), glutathione (GSH) and superoxide dismutase (SOD) levels. The most effective protection against the oxidative damage, with cell viability of 100%, was provided by 50 µg mL-1 of CNC-g-PNIPAM synthesized with 1:1 CNC:NIPAM mass ratio. Furthermore, there was no significant difference between the levels of GSH, MDA and SOD between cells treated with the modified CNC and cells not exposed to peroxide. These results indicate that the antioxidant properties of CNC-g-PNIPAM may be useful in possible applications in health treatments. INTRODUCTION Cellulose is the most abundant polymer in nature and consists of the union between monomers of D-glucopyranose through β-1,4 glycosidic bonds. It can be obtained from renewable sources such as hemp, linen, jute, ramie, cotton, palm, and coconut. Cellulose nanocrystals (CNCs) are crystalline domains of cellulose isolated from acid or enzymatic hydrolysis, with at least one dimension smaller than 100 nm.1-3 Cellulose nanocrystals have been widely investigated due to their special surface chemistry, environmental biodegradability, biocompatibility, low toxicity, and non-abrasive nature. The presence of hydroxyl groups (-OH) on CNC surfaces allows many types of chemical modifications and enables the production of materials with a wide range of properties and functions.4,5 Thermoresponsive character can be introduced to cellulose nanocrystals through grafting reactions with poly(N-isopropylacrylamide) (PNIPAM). This polymer responds through changes in its solvation state after heating or cooling around a critical temperature, 32 ºC, known as its lower critical solution temperature (LCST).6,7 Due to these unique properties, the combination of CNC with PNIPAM through grafting polymerization would be of great interest for various biomedical applications, such as drug delivery systems, contact lenses, wound dressing, scaffolds for tissue engineering, biosensing and cell encapsulation. Recently, PNIPAM-CNC hybrid hydrogels provided slow and sustained release of metronidazole, at 37 ºC.8 In the same trend, fluorescent and thermoresponsive CNC-g-(N-isopropylacrylamide-co-4-ethoxy-9-allyl-1,8-naphthalimide) particles exhibited pH and temperature dependence on the release of 5-fluoracil.9,10 In another study, the in vitro toxicity of CNC-g-PNIPAM using MTT and LDH assays was evaluated on two different cell lines (J774A.1 and MCF-7). The results indicated that cells had no major changes in cell viability, membrane permeability and cell morphology.11 Antioxidant materials are substances that slow down the rate of oxidation through one or more mechanisms, such as free radical inhibition.12 Many papers have shown cellulose nanocrystals and their derivatives as antioxidant agents, for example, in food packaging to extend its expiration date.13,14 CNCs can also have an antioxidant effect indirectly, for example, β-cyclodextrin-grafted CNCs increased the activity of superoxide dismutase, which in turn acts as an antioxidant agent in the intracellular environment.15 The results indicated that, when modified, CNCs can protect cell lines and other compounds from oxidative stress. In this paper, three CNC-g-PNIPAM copolymers were synthesized using free radicals as the initiation system at different CNC/NIPAM feed ratios. In addition, the cytotoxicity and antioxidant activity were evaluated to know the potential applicability in biomedical devices.
EXPERIMENTAL Materials Cotton fibers were obtained from York and employed as the cellulose source. N-isopropylacrylamide (NIPAM), N,N',N'-tetramethylethylenediamine (TEMED), 3-(4,5-dimethylthiazol-2-yl)-2-5-diphenyltetrazolium bromide (MTT), penicillin, streptomycin, dithiobisnitrobenzoic acid (DTNB), Griess reagent, hydrogen peroxide (H2O2), 4',6-diamidino-2-phenylindole (DAPI), paraformaldehyde and Triton X-100 were supplied from Sigma-Aldrich. Potassium persulphate (KPS) and trichloroacetic acid were purchased from Vetec. Sulfuric acid (H2SO4) and hydrochloric acid (HCl) were obtained from Synth. 1-Methyl-2-phenylindole and acetonitrile were acquired from Merck. Culture media components (minimum essential Dulbecco's modified Eagle medium - DMEM), fetal bovine serum (FBS) and phosphate buffered saline (PBS) were purchased from Invitrogen Corporation. Superoxide dimutase (SOD) activity kit was supplied by Enzo Life Sciences. Crystal violet was obtained from Electron Microscopy Science. Murine macrophage cell line (RAW 264.7 ATCC® TIB-71TM) was obtained from the Banco de Células do Rio de Janeiro (BCRJ). Preparation of CNC-g-PNIPAM Cellulose nanocrystals (CNCs) were prepared and characterized as described previously by our research group, but with some modifications.16 In this case, PNIPAM chains were grafted from CNCs by using the KPS/TEMED redox initiation pair (Table 1), under N2(g) atmosphere, at room temperature (~ 25 ºC), for 3 h, under constant mechanical stirring. The materials were purified by dialysis against water and lyophilized.
Nuclear magnetic resonance spectroscopy Carbon-13 nuclear magnetic resonance spectroscopy (13C NMR, Bruker Avance III HD 600 MHz) was performed with the samples in solid state, by combining cross-polarization with magic angle spinning (CP/MAS), spinning speed of 10 kHz, acquisition time of 30 ms and contact time of 2 ms. Cell culture The RAW 264.7 ATCC® TIB-71TM cell lines were cultivated in Dulbecco's modified Eagle's medium (DMEM) with 10% of fetal bovine serum, 10 mg mL-1 streptomycin and 10,000 IU penicillin. The cells were maintained at 5% CO2 and 37 ºC. MTT citotoxicity assay The effect of CNC, PNIPAM and modified CNCs on cell viability was evaluated by the ability to reduce formazan from 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) by mitochondrial enzymes in cells with active metabolism.17 The 1 × 104 cells (RAW 264.7 ATCC® TIB-71TM) were incubated in the presence of: (i) DMEM medium + 10% FBS (control: MTT reduction = 100%); (ii) DMEM + 10% FBS with CNC (75 mg mL-1); (iii) DMEM + 10% FBS with PNIPAM (75 mg mL-1) and (iv) DMEM + 10% FBS with modified CNCs (10; 25; 50; 75; 100 mg mL-1), at 37 ºC, for 24 h. Then, the medium was removed and the MTT was added. After 4 h in 5% CO2 at 37 ºC, the MTT was aspirated, and the formazan crystals were dissolved in 96% ethanol. Absorbance was measured at 570 nm18 in an Epoch microplate spectrophotometer. Proliferation assay Cell proliferation was determined by crystal violet assay.19 RAW 264.7 cells (15 × 103 cells well-1) were exposed or not (control group) to NCC (75 µg mL-1), PNIPAM (75 µg mL-1) and the modified CNCs (10, 25, 50, 75 and 100 µg mL-1), for 72 h. The resulting absorbance was read in an Epoch microplate spectrophotometer at 570 nm. Induced oxidative stress assay In order to test the protective capacity of the polymers against oxidative damage on RAW 264.7 cells caused by H2O2, a method described previously20 was performed as follows. First, optimum conditions for maximum cell damage were determined. RAW 264.7 cells were placed in 96 well plates (4 × 103 cells well-1) containing Dulbecco's modified Eagle medium (DMEM) without fetal bovine serum (FBS) and with different concentrations (0.1 to 5 mmol L-1) of hydrogen peroxide (H2O2), for 1 h. Afterward, the H2O2-containing medium was replaced with DMEM containing FBS. After 24 h of incubation, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide colorimetric (MTT) reduction ability of cells was evaluated. Here, the presence of H2O2 (3 mmol L-1) caused the cells to suffer enough damage to decrease their ability to reduce MTT about 60% (data not shown). Therefore, cells (4 × 103 cells per well) were incubated in a serum-free medium for 24 h. The medium was then exchanged with serum-containing the polymers and 3 mmol L-1 H2O2. The cells were further incubated for 2 h in this medium. Then, the medium was exchanged with serum, and cells were incubated for another 24 h. Finally, cell viability was determined by using the MTT test.18 DMEM with H2O2 (3 mmol L-1) and with serum were used for positive and negative controls, respectively. Nuclear morphology The RAW 264.7 cells were subjected to the experimental conditions described in the previous section. After, the cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 30 min, at room temperature. After washing twice with PBS, cells were maintained in PBS containing 0.1% Triton X-100, at room temperature, for 30 min. Fixed cells were washed with PBS and stained with DAPI (4',6-diamidino-2-phenylindole) (1 μg mL-1) solution for 30 min, at room temperature. Nuclear morphology of cells nuclei was examined under a fluorescent microscope (TE-Eclipse 300, Nikon). Data presented are representative of those obtained in at least three independent experiments carried out. Superoxide dismutase (SOD) evaluation The superoxide dismutase (SOD) activity is based on the ability of SOD to neutralize superoxide ions created by the xanthine/xanthine oxidase system and subsequently inhibits the reduction of WST-1 (water soluble tetrazolium salt) to WST-1 formazan. Briefly, RAW cells (5 × 106 in six-well plate), obtained 24 h after oxidative stress induction, were washed with ice-cold 1× PBS, and lysed as described in SOD activity kit protocol. The supernatant of each sample was collected, and the total SOD activity was assayed spectrophotometrically at 450 nm in an Epoch microplate spectrophotometer. The SOD concentration, expressed in units per milligram of protein, was determined using the SOD standard curve. Glutathione evaluation To assess the total level of glutathione, RAW cells (5 × 106 in six-well plate), obtained 24 h after oxidative stress induction, were sequentially washed with ice-cold 1× PBS, removed, resuspended in PBS and centrifuged (3000× g at 4 ºC) twice for 5 min. After this process, the suspension obtained was then diluted (1:1) in 50% trichloroacetic acid and centrifuged during 15 min (3000× g at 4 ºC). After, cell supernatant was diluted with the same volume of 0.4 mol L-1 Tris buffer contained 0.01 mol L-1 dithiobisnitrobenzoic. The material was read at 412 nm in an Epoch microplate spectrophotometer. The results were expressed as nmol 10-6 cells. Malonaldehyde levels To assess lipid peroxidation, malonaldehyde (MDA) production was measured with thiobarbituric acid reaction. Briefly, the cells under the same conditions as described above were triturated in 20 mmol L-1 Tris-HCl buffer and centrifuged during 15 min (3000× g at 4 ºC). The chromogenic reagent (10.3 mmol L-1 1-methyl-2-phenylindole in acetonitrile, 3:1 v/v), and a 37% solution of HCl were dropped to each supernatant sample (150 mL). The samples were kept at 45 ºC, for 40 min, and then were centrifuged (15 min; 3000× g; 4 ºC). The absorbance was measured at 586 nm in an Epoch microplate spectrophotometer, and the results were expressed as nmol 10-6 cells. Immunomodulatory activity To immunomodulatory activity was evaluated by nitric oxide (NO) production assay on macrophages incubated with the samples. After 24 h of incubation for cell adhesion, the medium was removed and a new medium containing 50 µg mL-1 of CNC, PNIPAM and CNC-g-PNIPAM was added. After 24 h, the supernatant (100 µL) from each well was removed and added to 100 µL of Griess reagent (1% sulfanilamide in 5% phosphoric acid and 0.1% naphthylethylenediamine dihydrochloride in water). The presence of nitric oxide was observed by color change, which was monitored using a microplate spectrophotometer (Epoch; BioTek, Winooski, VT, USA) at 560 nm. For control, it was used the medium from RAW cells or the samples. Sodium nitrite was employed as the standard. Statistical analysis The data were expressed as the mean ± standard deviation. Statistical analysis was performed using a simple variation analysis (one-way ANOVA) followed by the Bonferroni test (p < 0.05). Statistical analysis were performed using the GraphPadPrism® software, version 5.0 (GraphPad).
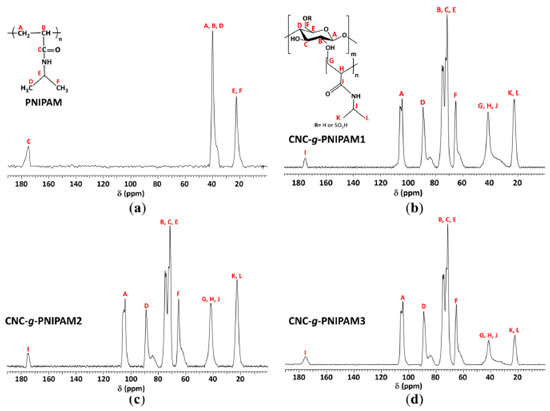
RESULTS AND DISCUSSION 13C NMR spectroscopy The modified nanocrystals CNC-g-PNIPAM were prepared by means of the grafting from route, using the TEMED/KPS redox pair as free-radical initiators. The confirmation of grafting was indicated by 13C NMR spectra (Figure 1). They presented peaks related to PNIPAM repeat units: isopropyl groups at 22 ppm (A and B); C, E and F carbons at 41 ppm; and a downfield peak at 175 ppm, ascribed to C=O (D).21 The spectra of the copolymers also exhibited peaks related to the cellulose structure: at 65 ppm due to hydroxymethyl F carbons; a signal centred at 75 ppm, due to B, C, and E carbons; a peak at 89 ppm, due to C4 carbon; and a signal at 105 ppm, assigned to A carbon. Besides, the absence of signals at 120-130 ppm (C=C) indicates that the products are free from monomer impurities and the purification step was efficient.22,23
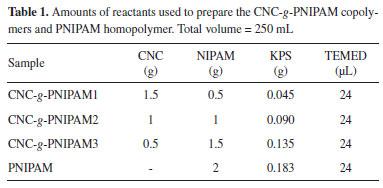
MTT and proliferation assays Initially, we would like to assess the ability of the synthesized materials to protect cells from oxidative stress. However, it was necessary to rule out the possibility that they are toxic for RAW cells. So, we first performed cytotoxicity and proliferation assays on RAW 264.7 as the macrophage cells. The results of 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assays are shown in Figure 2(a). The results were expressed as a percentage of MTT reduction. The negative control consisted of medium with serum and was used as reference to determine the value corresponding to 100%.
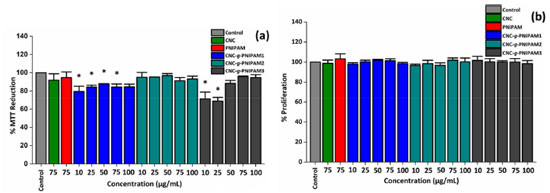 Figure 2. (a) Reduction of MTT to formazan (%) by cells exposed to homopolymer PNIPAM, unmodified CNC and modified CNC-g-PNIPAM after 24 h of treatment; * indicates significant difference between the control and samples (p < 0.05). Also, (b) cell proliferation assay performed by the crystal violet technique measuring cell viability. The experiment was carried out in RAW cells untreated (control) and treated with the synthesized materials
Neither CNC nor PNIPAM alone affected the ability of RAW cells to reduce MTT. In addition, CNC-g-PNIPAM2, regardless of the added concentration, also did not affect the ability to RAW cells to reduce MTT. These results indicate that these polymeric materials, at the evaluated concentrations, have no cytotoxic effect against RAW cells. The cells treated with CNC-g-PNIPAM3 (10 and 25 µg mL-1) decreased their ability to reduce MTT at about 30%. However, this effect was abolished when cells were exposed to higher concentrations (from 50 to 100 µg mL-1) of the compound. When cells were exposed to CNC-g-PNIPAM1, regardless of the concentration evaluated, they showed a decrease of approximately 20% in their ability to reduce MTT. These data indicated that the synthesis conditions of the compounds can affect the cellular metabolism. The effect of these compounds on RAW cell viability was also evaluated. As shown in Figure 2(b), the compounds, regardless of the evaluated condition, did not affect cell viability. This indicates that although CNC-g-PNIPAM1 and CNC-g-PNIPAM3 affect the ability of cells to reduce MTT, they do not decrease RAW cell viability. To compare the action of the compounds on cells exposed to stress conditions, the concentration of 50 mg mL-1 was chosen to be used in next tests, since at this concentration most compounds did not show a cytotoxic effect. Effect of CNC-g-PNIPAM on viability of RAW cells exposed to hydrogen peroxide (H2O2) A thermoresponsive gel containing PNIPAM was synthesized by Yang et al.24 and these authors showed that this gel had an intrinsic antioxidant property in several in vitro tests. Furthermore, these authors suggested that its antioxidant activity comes from PNIPAM. Taking it a step further to demonstrate the potential of PNIPAM as an antioxidant biomaterial, herein the antioxidant activity of PNIPAM as well as CNC-g-PNIPAM1, CNC-g-PNIPAM2 and CNC-g-PNIPAM3 was evaluated in cells exposed to oxidative damage caused by H2O2. In Figure 3, the effect of hydrogen peroxide (H2O2) on the ability of cells to reduce MTT is shown. In contrast to the control, the cells exposed only to H2O2 were able to reduce just 37% of MTT. The tested compounds had different actions. The cells exposed to CNC reduced MTT by approximately 80% compared to the control cells. Whereas the cells exposed to CNC-g-PNIPAM2 reduced MTT by approximately 95% when compared to the control cells. CNC-g-PNIPAM1 was not able to protect cells from the action of H2O2. In addition, CNC-g-PNIPAM3 showed a low protective effect, since the cells reduced MTT by approximately 50%.
 Figure 3. MTT reducing activity of the RAW cells incubated with CNC, PNIPAM and CNC-g-PNIPAM (50 µg mL-1) and hydrogen peroxide (H2O2) for 24 h. Values are expressed as the mean ± standard deviation. * Indicates the significant difference between the control and samples (p < 0.05). aindicates significant difference between the "control + peroxide" and the tested samples (p < 0.05)
Among samples tested, the most effective antioxidant was CNC-g-PNIPAM2. The percentage of MTT reduction obtained for modified CNC-g-PNIPAM2 was higher than for the unmodified CNC. When cells were exposed to CNC-g-PNIPAM2 (50 µg mL-1), their ability to reduce MTT was three times higher than that observed for the cells treated with peroxide (control + peroxide). To confirm the effect of CNC-g-PNIPAM2 in the MTT tests, RAW macrophage cells were exposed to stress conditions in the presence of all modified materials, and nuclear DAPI staining was evaluated. As shown in Figure 4, nuclei with condensed chromatin of different sizes, containing well-preserved but compacted cytoplasmic organelles and/or nuclear fragments, were observed in H2O2 treated RAW cells. The findings showed that CNC-g-PNIPAM2 protects cell nuclei from damage caused by hydrogen peroxide.
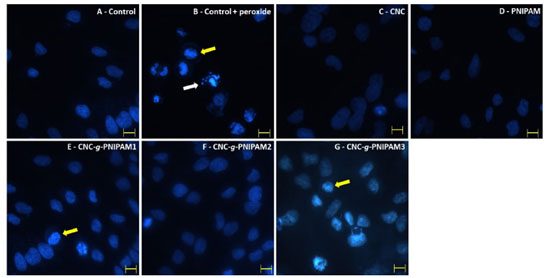 Figure 4. Nuclear morphology of : (a) untreated cells; (b) cells treated with H2O2; cells treated with 50 µg mL-1 of materials: (c) CNC; (d) PNIPAM; (e) CNC-g-PNIPAM1; (f) CNC-g-PNIPAM2; (g) CNC-g-PNIPAM3. White arrows indicate nuclear fragmentation and yellow arrows indicate chromatin condensation. Magnification × 400 bars correspond to 15 μm
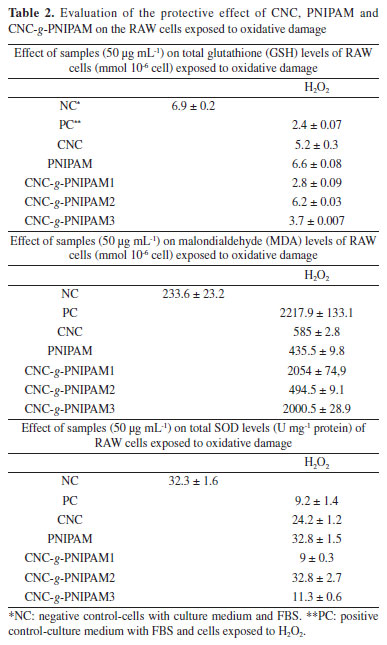
To further confirm that the oxidative stress in the cells was reduced by the presence of CNC-g-PNIPAM2, we evaluated the levels of three markers of oxidative stress SOD, GSH, and malondialdehyde (Table 2). Free radical-mediated cell damage can generally cause lipid peroxidation. Malondialdehyde level is a good oxidative marker, as the greater the amount of this molecule, the greater the lipid peroxidation. The presence of PNIPAM and CNC-g-PNIPAM2 prevented malondialdehyde levels from getting so high. When cells are exposed to oxidative damage, the concentration of SOD and GSH decreases considerably. This is because SOD and GSH are consumed by the reactive species that are formed. When RAW macrophage cells are exposed to oxidative damage, there is a decrease in the levels of SOD and GSH. In both cases, it was observed that the modified CNC-g-PNIPAM2 protected the cells against the oxidative damage.
CNC-g-PNIPAM1 and CNC-g-PNIPAM3 have been synthesized with the same components as CNC-g-PNIPAM2. However, CNC-g-PNIPAM2 showed much better antioxidant activity. The difference between these three products is that during their synthesis, the proportion of CNC and NIPAM was different. This affected their antioxidant activity. In another paper, the preparation of α-tocopherol loaded nanoparticles (TOC-NP) based on amphiphilic thermosensitive triblock copolymers containing PNIPAM was reported.25 In that case, the increase or decrease of NIPAM amount in the synthesis medium did not influence the antioxidant activity. This demonstrates that addition of PNIPAM to a conjugate will not necessarily provide antioxidant activity to that one, and there must be a small range in which the amount of NIPAM added to the new material can transfer its antioxidant properties. Through grafting chitosan oligosaccharide on CNC, the resulting product showed antioxidant properties.26 The authors suggested that the high in vitro antioxidant activity of the modified CNC compared to its precursors was due to the orientation that the functional groups of CNC and chondroitin assumed after the conjugation process, leading to increased accessibility of the functional groups to interact to reactive species. Therefore, it seems clear that the amount of PNIPAM and other components that were used in synthesis of CNC derivative influences how their functional groups are exposed and accessible, which in turn, provides or not their properties to the new CNC derivative. Therefore, this would explain the fact that CNC-g-PNIPAM2 is the compound with greater antioxidant activity than the other CNC derivatives studied here. Evaluation of nitric oxide release by RAW cells exposed to compounds Many papers show that macrophages can be stimulated and become active as agents of the inflammatory process. The inflammatory process is often unwanted, as it is harmful to organisms. Therefore, the pro-inflammatory potential of the compounds studied here was evaluated by the levels of nitric oxide released by RAW cells after being exposed to these compounds. We identified that no material was able to significantly affect the release of NO by macrophages (data not shown). We can consider the materials as inert in this case, that is, they did not attack and did not affect their cell growth.
CONCLUSIONS In this paper, it is presented the synthesis of CNCs decorated with PNIPAM chains, via free-radical polymerization. The success of the grafting was confirmed through 13C NMR spectra. The modified CNCs did not either affect the cell viability (RAW 264.7 cell line) or the RAW cell oxide nitric production. CNC-g-PNIPAM2 presented the highest antioxidant activity among all copolymers tested. They also exhibited better antioxidant activity when compared to their precursors PNIPAM and CNC. In summary, the 1:1 CNC/NIPAM mass ratio used in the reaction system (CNC-g-PNIPAM2) stood out over the other conditions employed leading to a product of improved antioxidant activity. In addition, CNC-g-PNIPAM2 were able to protect RAW cells from the oxidative stress caused by H2O2.
ACKNOWLEDGMENTS This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES - finance code 001). The authors are grateful to PETROBRAS S/A (SAP 4600580022 and 4600568098) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grants 304339/2019-9 and 309771/2019-6) for their financial support. The authors are also grateful to the Laboratório de Espectroscopia e Polímeros (LEPOL) from UFSM for the NMR analysis.
REFERENCES 1. Börjesson, M.; Westman, G. In Cellulose - Fundamental Aspects and Current Trends; Poletto, M.; Ornaghi Junior, H. L. , eds.; IntechOpen: London, 2005. [Crossref] 2. Nascimento, D. M.; Nunes, Y. L.; Figueirêdo, M. C. B.; de Azeredo, H. M. C.; Aouada, F. A.; Feitosa, J. P. A.; Rosa, M. F.; Dufresne, A.; Green Chem. 2018, 20, 2428. [Crossref] 3. Silva, D. J.; D'Almeida, M. L. O.; O Papel 2009, 70, 34. [Link] accessed in August 2023 4. Grishkewich, N.; Mohammed, N.; Tang, J.; Tam, K. C.; Curr. Opin. Colloid Interface Sci. 2017, 29, 32. [Crossref] 5. Kontturi, E.; Laaksonen, P.; Linder, M. B.; Nonappa; Gröschel, A. H.; Rojas, O. J.; Ikkala, O.; Adv. Mater. 2018, 30, 1703779. [Crossref] 6. Haqani, M.; Roghani-Mamaqani, H.; Salami-Kalajahi, M.; Cellulose 2017, 24, 2241. [Crossref] 7. Yang, L.; Fan, X.; Zhang, J.; Ju, J.; Polymers 2020, 12, 389. [Crossref] 8. Zubik, K.; Singhsa, P.; Wang, Y.; Manuspiya, H.; Narain, R.; Polymers 2017, 9, 1. [Crossref] 9. Wu, W.; Huang, F.; Pan, S.; Mu, W.; Meng, X.; Yang, H.; Xu, Z.; Ragauskas, A. J.; Deng, Y.; J. Mater. Chem. A 2015, 3, 1995. [Crossref] 10. Wu, W.; Li, J.; Liu, W.; Deng, Y.; BioResources 2016, 11, 7026. [Crossref] 11. Jimenez, A.; Jaramillo, F.; Hemraz, U.; Boluk, Y.; Ckless, K.; Sunasee, R.; Nanotechnol. , Sci. Appl. 2017, 2017, 123. [Crossref] 12. Nimse, S. B.; Pal, D.; RSC Adv. 2015, 5, 27986. [Crossref] 13. Criado, P.; Fraschini, C.; Salmieri, S.; Becher, D.; Safrany, A.; Lacroix, M.; Ind. Biotechnol. 2015, 11, 59. [Crossref] 14. Criado, P.; Fraschini, C.; Jamshidian, M.; Salmieri, S.; Safrany, A.; Lacroix, M.; Cellulose 2017, 2, 2111. [Crossref] 15. Sunasee, R.; Carson, M.; Despres, H. W.; Pacherille, A.; Nunez, K. D.; Ckless, K.; J. Nanomater. 2019, 2019, 4751827. [Crossref] 16. Pinheiro, J. A.; Marques, N. D. N.; Villetti, M. A.; Balaban, R. D. C.; Int. J. Mol. Sci. 2020, 22, 352. [Crossref] 17. Mosmann, T.; J. Immunol. Methods 1983, 65, 55. [Crossref] 18. Amorim, M. O. R.; Gomes, D. L.; Dantas, L. A.; Viana, R. L. S.; Chiquetti, S. C.; Almeida-Lima, J.; Costa, L. S.; Rocha, H. A. O.; Int. J. Biol. Macromol. 2016, 93, 57. [Crossref] 19. Gillies, R. J.; Didier, N.; Denton, M.; Anal. Biochem. 1986, 159, 109. [Crossref] 20. Fernandes-Negreiros, M. M.; Batista, L. A. N. C.; Viana, R. L. S.; Sabry, D. A.; Paiva, A. A. O.; Paiva, W. S.; Machado, R. I. A.; de Sousa Junior, F. L.; Pontes, D. L.; Vitoriano, J. O.; Alves Junior, C.; Sassaki, G. L.; Rocha, H. A. O.; Antioxidants 2020, 9, 1. [Crossref] 21. Gao, X.; Cao, Y.; Song, X.; Zhang, Z.; Xiao, C.; He, C.; Chen, X.; J. Mater. Chem. B 2013, 1, 5578. [Crossref] 22. Wang, M.; Ge, H.; Song, J.; Xu, M.; Chem. J. Chin. Univ. 2015, 36, 1422. [Crossref] 23. Zoppe, J. O.; Habibi, Y.; Rojas, O. J.; Venditti, R. A.; Johansson, L.; Efimenko, K.; Österberg, M.; Laine, J.; Biomacromolecules 2010, 11, 2683. [Crossref] 24. Yang, J.; van Lith, R.; Baler, K.; Hoshi, R. A.; Ameer, G. A.; Biomacromolecules 2014, 15, 3942. [Crossref] 25. Quintero, C.; Vera, R.; Perez, L. D.; Polimeros 2016, 26, 304. [Crossref] 26. Akhlaghi, S. P.; Berry, R. M.; Tam, K. C.; AAPS PharmSciTech 2015, 16, 306. [Crossref] |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access