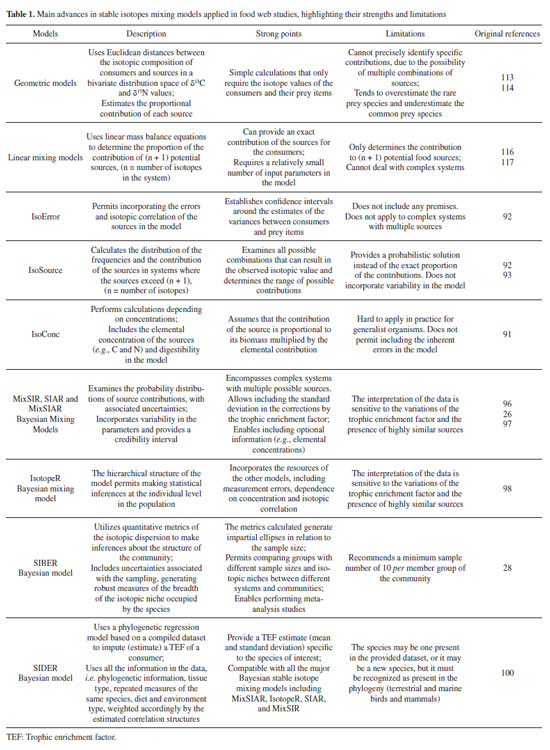
Revisão
| Stable isotopes and mercury as tools to depict aquatic food webs |
|
Cynara Pedrosa FragosoI; Pedro Vianna GattsI; Ana Paula Madeira Di BenedittoI; Luiz Antonio MartinelliII; Luiz Drude de LacerdaIII; Carlos Eduardo de RezendeI*
I. Centro de Biociências e Biotecnologia, Universidade Estadual do Norte Fluminense Darcy Ribeiro, 28013-602 Campos dos Goytacazes - RJ, Brasil Recebido em 23/05/2023; *e-mail: crezende@uenf.br The structure of food webs is a fundamental attribute of ecosystems, and their characterization provides an intrinsic knowledge of the trophic interactions among organisms and of nutrient and energy transfer within ecosystems. Over the last few decades, several chemical and biogeochemical approaches have been proposed to explore different aspects of food webs. In this sense, this study reviews the application of stable isotopes and mercury as main auxiliary tools to the characterization and modeling of aquatic food webs, including analytical and modeling advances, strengths and limitations. The metanalysis performed showed that the most used tools for trophic ecology studies are stable isotopes, and that they can provide better results when combined with mercury and specific chemical molecules such as amino and fatty acids. In addition, the statistical methods applied in the interpretation of results, such as isotope mixing models, have witnessed significant advances in the last two decades. All approaches have premises and limitations when applied to aquatic ecosystems, which must be well understood prior to results interpretation. The use of multiple tracers in trophic studies provides complementary information and in many cases is an appropriate alternative to overcome some limitations, allowing to expand the knowledge of the food webs. INTRODUCTION Food webs are networks of interactions between consumers and their nutritional sources, encompassing models of flow of nutrients, matter, and energy through ecosystems.1 They include populations, groups of organisms, or trophic units, which constitute the primary bases for the development and advancement of ecological studies.2,3 Since the initial studies of food chains, important concepts have been introduced, allowing a better understanding of energy flow, estimates of primary production, and trophic relationships.4,5 A historical summary of major studies on the ecology of food webs can be found in Layman et al.3 The current knowledge about the energy flow in ecosystems benefited from using new tools like stable isotope analysis.6,7 Stable isotopes are non-radioactive nuclides of the same chemical element (same number of protons) but with a different number of neutrons, resulting in different mass numbers.8-10 Stable isotope analysis (SIA) has a wide range of applications in geochemistry and biogeochemistry,11-14 and has become a highly effective tool for tracing the putative food sources in complex food webs (e.g., habitats, types of resources) and determining the consumer's trophic position. Moreover, it provides in time and space integrated information on the complex interactions between organisms, permitting the development of models to represent the structure of trophic webs in nature.7,15 The isotope ratios most commonly used as tracers in food webs are carbon (13C/12C) and nitrogen (15N/14N).9,10 Additionally, isotope ratios of sulfur (34S /32S) are increasingly being used in trophic studies.16-18 In combination with SIA, quantifying mercury (Hg) concentrations, an element that suffers enrichment through the food chain, a processes called biomagnification, helped model food webs.19,20 Another promising approach is associating SIA of specific components with studies of amino acids and fatty acids to expand the possible applications to the field of trophic ecology.7,21-23 In the past three decades, many researchers have shifted from the qualitative focus of the pioneer isotopic applications to more quantitative data analyses to improve the knowledge on food webs.7,24,25 These advances have given rise to the need for improved statistical analysis and of modelling to enable a more robust study of SIA.7,25-29 The development of new approaches for analyzing isotope ratios has prompted questions about which method or model is most suitable for a particular purpose. This entails recognizing the strengths and uncertainties of each method and its applications depending on the circumstances and specific questions addressed by a given study. Therefore, this review has two objectives: (i) describe the use of stable isotopes as well as mercury as the main accessory tools used to model aquatic food webs and their applications; and (ii) summarize the recent advances of the main statistical models employed in studies of trophic ecology, identifying their strength, limitations, and future perspectives. Stable isotopes Isotope ratios express the difference between the number of neutrons in the nucleus of heavier and lighter stable isotopes of the same element. For example, the atomic species 13C has one more neutron in its nucleus than its more abundant form, 12C. These are called heavy and light carbon isotopes in the literature.6,9,10 The isotope values are generally reported by the delta (δ) notation where δ13C denotes the ratio of the heavy and light carbon isotopes of the sample compared to an international standard: Vienna Pee Dee Belemnite (VPDB), according to the following Equation 1: 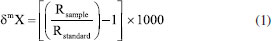 where X is the chemical element to be analyzed, m is the mass of the heaviest isotope and R is the ratio between the heaviest and lightest isotope, both in the sample and the standard. The final value is expressed in parts per thousand (%).6,9 DeNiro and Epstein8 were the first to show that the isotopic values of consumers reflected the isotopic composition of the ingested food item, discounting eventual small isotopic discrimination between sources and consumers. Since then, isotopes have been used as chemical markers in biological tissues to determine not only the ingested food but, more importantly, the portion of it finally assimilated.9 This supplies information about trophic interactions, energy transfer, and food web complexity, as well as identifying temporal or spatial alterations of major resources and consumers' diets.7,25 Based on isotopic data alone, a clear constrain of this approach is the incapacity to identify the food items ingested with the same accuracy obtained by analyzing the stomach contents or the direct observation of the foraging behavior.30 Additionally, estimating the biomass of items consumed only by differentiating the feeding pattern and the trophic level is impossible.7 This approach also needs previous knowledge of the life history of the target species,31,32 and of the isotopic baseline values of food items consumed; the frequent lack of such information is a serious shortcoming for estimation of the ecosystem's trophic structure. Thus, isotopic tools can be complementary to the information acquired via traditional observation methods mentioned.9 In the past two decades, studies associating stable isotopes with organisms' diets, trophic relations, and aquatic food webs significantly increased (Figure 1), and various previous reviews have also reported a same trend of an increasing number of studies associating isotopic analysis with organisms' diets, trophic relations, and the food web.7,25,33
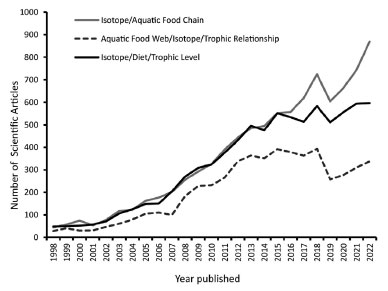 Figure 1. Number of papers published in the past (~ 20 years) employing stable isotopes in aquatic food webs, cataloged by the CAPES/MEC Journals Portal. The keywords used were "isotope", "aquatic food chain", "aquatic food web", "diet" and "trophic relationships"
Some premises should be considered to enable the proper application of isotope ratios in trophic ecology studies, such as (i) the groups of primary producers considered as baseline food sources for any food web (e.g., phytoplankton, algae, plant material) typically have distinct isotope values, permitting tracing the origin of the carbon quantified in higher trophic levels; (ii) the buildup of the tissues of the consumers reflects the isotopic composition of their diets; and (iii) the conservative character of isotopes ratios allows identifying a constant and predictable variation of their values among the different components of the food web, a process known as isotopic fractionation or the trophic enrichment factor (TEF).10,34,35 A noteworthy caveat regards the second premise, since many factors can affect the isotopic assimilation of a particular food item in the tissues of organisms. Boecklen et al.35 listed biotic and abiotic factors that are sources of variation in the isotopic composition of organisms, ranging from the properties of consumers (e.g., types of tissue, life histories, physiological conditions), properties of the ingested food (e.g., lipids and proteins contents, elemental concentration), environmental properties (e.g., biome and habitat type) and analytical properties (e.g., capacity of lipids extraction, precision and reproducibility of methods). Indeed, TEF is one of the central matters for adequate interpretation of the energy flow, inter-relationships, and complexity of the food web.9,27,33 The δ13C isotopic composition allows identifying which primary producers or organic sources form the baseline of a given consumer since the isotopic composition of producers is assimilated in the tissue of consumers with a TEF generally of ≤ 1% at each trophic level, the so-called fractionation.15,36 Conversely, the isotopes of δ15N allow evaluation of an organism's trophic position in the food chain. Each advance of the trophic level generally involves losses of the lightest isotope (14N) and retention of the heaviest one (15N), resulting in an increase in the value of δ15N along the trophic chain.6,37 Therefore, their ratios can be used to estimate the food chain's length.2 Each change in the trophic level is associated with an average increase of ~ 3% of δ15N. However, this value can vary significantly in function of the species, tissue analyzed, and temporal and spatial variations, with values ranging from ~ 2 to 5%.7,15,38 The potential applications of δ 13C and δ 15N are diverse, and consequently, numerous studies focused on trophic ecology have been conducted. These studies include determining the origin of fishing stocks,39 comparing trophic niches and positions,40 investigating trophic links and feeding areas,41 tracing individual niche trajectories by analyzing isotopic assessments of various body tissues,42 and identifying the sources of organic matter on the continental shelf, among others.43 The isotopic composition of sulfur, expressed as δ34S, is another tool increasingly applied in trophic studies.16,18,44,45 In aquatic ecosystems, the different chemical forms of S (e.g., sulfate in the water column, sulfides in the sediment) tend to have different values of δ34S. The reduction of aqueous SO42- by sulfate-reducing bacteria results typically in the deposition of sulfide compounds in the sediment, with lower values of δ34S. This difference is more pronounced in anoxic environments. As a result, primary producers that use sulfates from the water column tend to be more enriched in 34S (e.g., microalgae and phytoplankton, with average δ34S ~ 18%). In contrast, those that use the sulfides from the sediment are more depleted in 34S (e.g., plants in flooded areas, with δ34S between −10% and 5%).44 Since the isotopic fractionation of S is usually very low it is frequently disregarded, its values permit differentiating benthic and pelagic consumers in coastal habitats, according to the sources of primary production in the food web. This allows differentiation in environments where there is a possible overlap of organic matter sources.44,46 Moreover, the tool can also be applied to species that differ regarding habitat use, such as identifying movements between shallow and deep water or investigating migratory species.16,18 Two critical points should be considered in interpreting stable isotopes in organisms. The first is the turnover rate, which refers to renewing or synthesizing new biological tissue due to dietary alterations. It reflects the period necessary for the consumer to integrate the isotopic signal of its prey. Therefore, the isotope ratio of the consumer reflects the different feeding periods due to different turnover rates associated with the metabolism of each tissue.47,48 For example, tissues with higher metabolic activity (e.g., liver and blood plasma) have faster turnover rates, allowing detection of the recent diet (e.g., few days or hours) or in short experiments. Alternatively, tissues that have slower turnover rates, like muscles and bones, are better suited for research on medium and long-term effects.7,35 The turnover rate is also influenced by inherent developmental variations that impact metabolism, such as periods of rapid growth or food restriction that slow it down.49 When designing a sampling approach, it is crucial to collect consumers and their food sources at appropriate temporal and spatial scales aligning with the isotopic signal in their tissues. Typically, sources are collected before their corresponding consumers.7,25 The second point to be considered involves one of the premises of SIA, the trophic isotopic fractionation. Questions about the generalized use of the values widely applied in trophic ecology studies have been raised.8,15 Studies indicated that these values can vary significantly according to some physiological and environmental aspects, such as the type of ecosystem, type of tissue, diet, trophic position, taxonomic group, the form of excretion (ammonia, urea or uric acid) and treatment of samples (e.g., extraction of lipids).25,46,50 Therefore, the best choice is to apply, site-specific situation, generating more precise estimates for small groups of organisms or specific consumers (e.g., taxon, tissue, diet).25 The ideal design is to create values near the real ones in each case through brief experiments or observations. However, this alternative is only available on some occasions. In these cases, seeking more robust estimates in meta-analysis studies is possible, which use mathematical correction factors to minimize the inherent errors in fractionation values.33,38,46,51 Some studies have indicated the need for new advances and experimental studies seeking to understand the nature of the variations observed in TEF.7,25,52 Mercury Mercury (Hg) is a metal with high toxic potential and considered a global contaminant due to atmospheric circulation and deposition.53 High Hg levels cause adverse effects in ecosystems and the associated food webs, generating risks to human health.37,54 In aquatic environments, emissions of Hg that are buried in surface sediments into the water column can be a significant source of the metal for organisms.55 The bioavailability through methylation56-59 is one of the determinants of Hg contamination in coastal food webs. In the biota, methylmercury is the predominant form, possibly varying from ~ 80 to 90% of the total Hg in organisms, whereas in other components of food webs contains less proportions (sediments < 1%, suspended particulate matter ~ < 5%, seston ~ < 10%).60 Some inherent characteristics of this mercury species favor its retention in biological tissues, such as high permeability through cell membranes, an affinity for the sulfhydryl group of proteins, high gastrointestinal absorption, stability, lipid solubility, and slow excretion rate (taking ~ 2.8 times longer than the inorganic form).54,61 Once incorporated in the biota, Hg can be bioaccumulated (i.e., assimilated and retained in the biological tissues relative to the abiotic environment), and biomagnified (i.e., increasing concentrations in higher trophic levels of the food chain). Few metals have been recognized as able to undergo biomagnification, meaning rising concentrations at each trophic level in the food chain (e.g., detritivores < herbivores < omnivores < carnivores and piscivores, in particular).37,59,61-66 Due to the clear connection between the level of Hg and the trophic position of organisms, researchers have sought to integrate information about feeding habits and trophic position with biomagnification and isotopic levels.19,66-70 Since 15N enrichment typically indicates an increase in the trophic level, many authors have reported a positive and significant relationship between δ15N and Hg.19,20,66,68-70 Additionally, the δ15N values can also be used to calculate the trophic level of consumers, allowing the association of these data with the concentrations of Hg and the phenomenon of biomagnification.20,66 An example of that association is depicted in Figure 2.
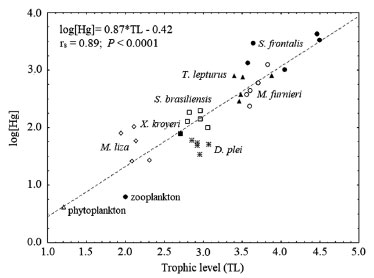 Figure 2. Relationship between the Hg concentration and trophic position of the organisms of a marine food web in Southeast Brazil (extracted from Kehrig et al.20 and reprinted with Criative Commons permission CC BYNC 4.0). X. kroyeri: Xiphopenaeus kroyeri (omnivore crustacean), D. plei: Doryteuthis plei (carnivore cephalopod), M. liza: Mugil liza (plankton feeder fish), S. brasiliensis: Sardinella brasiliensis (plankton feeder fish), M. furnieri: Micropogonias furnieri (benthic feeder fish), T. lepturus: Trichiurus lepturus (voracious predator fish), and S. frontalis: Stenella frontalis (voracious predator cetacean)
Another multiple application encompasses the concentrations of Hg and isotopes of C, N, and S in the trophic context.17,45 The variation of δ34S in coastal ecosystems is directly related to the activity of sulfate-reducing bacteria, which are considered to be mainly responsible for the methylation of Hg.17,56 Hence, multiple tracers can be used to estimate the biomagnification magnitude of Hg in the aquatic food webs and thus to investigate the anthropogenic impacts and the quality of the organisms more frequently consumed by humans.62,68,71 In recent decades, significant progress has been made in using stable isotopes of Hg to unravel connections between various components of aquatic food webs. Mercury possesses naturally occurring stable isotopes, each of which can undergo distinct fractionation patterns due to chemical, physical, or biological reactions, leading to both mass-dependent fractionation (MDF) denoted as δ202Hg, and mass-independent fractionations (MIF) resulting in odd-MIF represented as δ199Hg and δ201Hg, or even-MIF represented as δ200Hg and δ204Hg. The MDF δ202Hg has been used to define Hg sources in the environment due to different fractionation patterns imprinted by industrial or atmospheric processes, while Δ199Hg (MIF) has been used to trace the sources of mercury in marine food webs. Odd-MIF commonly results from photochemical transformations of Hg and δ199Hg values are useful for tracking the extent of Hg photochemical demthylation or reduction.72-74 Isotopic measurements have been widely applied in environmental and ecotoxicological studies focusing on Hg. They have found widespread use in food web investigations, helping to identify sources and trophic positions, as well as investigating dietary sources of methylmercury and its biomagnification.74 Typically, these measurements are combined with light stable isotopes (13C, 15N, 34S, and D) to resolve food webs in various contexts, such as: (i) differentiating between aquatic and terrestrial dietary sources in relation to varying Hg levels;75 (ii) estimating trophic positions to calculate trophic magnification slopes for methylmercury;37 and (iii) distinguishing between freshwater and marine-based food webs and their respective Hg levels.76 Advances have been made when coupling environmental ecological applications with food webs. This applications includes: (i) coastal and oceanic mixing of dietary with the former has more negative δ199Hg and δ202Hg values, suggesting the influence of exported Hg from rivers, the last had more positive values, like other oceanic areas, due Hg atmospheric precipitation coupled with oceanographic water mass transport;77,78 (ii) vertical profiling of animal migration with negative and/or lower δ199Hg, δ202Hg and δ13C values in higher depths compared to surface waters;74 (iii) vertical segregation between sea bottom in hydrothermal and cold seeps to water column, as δ199Hg values of methylmercury in the upper oceans are positive and decrease with depth and δ202Hg values of methylmercury are mainly positive, while those of inorganic Hg are predominantly negative in the upper oceans, there is little to no upper marine methylmercury in the chemosynthetic food webs of the deep sea;79 (iv) differences in source contamination when comparing isotopic compositions of total Hg and of methylmercury between different components of the food web, indicating distinct sources of Hg for sediments than for seston, invertebrates and fishes.60 Analysis of specific components - amino acids and fatty acids A significant technological evolution has expanded the possible applications of multiple tracers to investigate the structure of food webs and the quantification of specific compounds is likely the area with greatest potential for future development.7,21,23 The determination of the isotopic composition of specific amino acids (AAs) is a relatively new method carried out through isotopic analysis of nitrogen (δ15N), so called amino acid compound specific nitrogen isotope analysis (AA-CSIA). It is used to generate more precise estimates of the trophic position of organisms and to trace the baseline resources of a given food web in situations of uncertainties associated with traditional isotopic analysis.80,81 The technique is based on contrasting isotopic fractionation between two AA groups during metabolism. The first group is called "trophic" AA, represented by glutamic acid, aspartic acid, alanine, leucine, isoleucine, proline, and valine. The group's main representative is glutamic acid, which generally has greater trophic fractionation and is more easily identified by gas chromatography. The second group is "source" AA, represented by methionine, tyrosine, and phenylalanine. Among these, phenylalanine is most often used as a marker due to its lower variation of δ15N values during trophic transfer.82,83 During the metabolism of AAs, glutamic acid undergoes a deamination and transamination process, causing significant enrichment of δ15N at each trophic level (average fractionation of 8.0%). On the other hand, phenylalanine maintains its amine group during metabolism, since it cannot be synthesized by the organism itself. Therefore, the trophic enrichment of phenylalanine is substantially lower (average of 0.4%).21,82 As a result, the isotopic analysis of the "source" AAs in consumers provides information about the baseline resources of the food web assimilated in their tissues, due to the small fractionation value between trophic levels. On the other hand, the isotopic analysis of the "trophic" AAs reveals the trophic positioning of organisms within the food web due to the consistent and relatively large increase of the isotope values during trophic transfer.80,84 Compared with traditional SIA, the application of AA-CSIA can be a good alternative when limits exist regarding precise estimates of the baseline resource of a given food chain, mainly in aquatic systems or when the potential sources have very similar isotope values. The main analytic advantage of AA-CSIA is that analysis of a single consumer can provide information on the isotopic composition at the base of the chain and the number of trophic transfers.21,82,85 In some cases, this also eliminates the need to analyze primary producers, meaning that fewer samples are necessary to characterize consumers at higher levels of the food chain. Nevertheless, the cost of AA-CSIA is generally higher than that of traditional analysis. Another tool that can be applied to trophic ecology investigations is the analysis of fatty acids, which are carboxylic acids formed by long hydrocarbon chains that compose the lipids necessary for a range of metabolic functions. When consumed, they can be used as an energy source or be assimilated in the adipose tissue of the consumer (mainly in the form of triglycerol). This permits the fatty acid profiles from preys to be stored in the tissues of consumers in a predictable way.86,87 In this sense, the joint approach with isotopic analyzes provides complementary information in trophic studies and has been especially applied in marine ecosystems22,23,86 and estuaries.88 Some studies have combined analysis of stable isotopes, Hg, and essential fatty acids to characterize food webs in marine habitats and to improve the selection of trophic tracers to future studies.23,89 Three fatty acids are recognized as being essential to consumers and can be applied in trophic studies: docosahexaenoic acid (DHA), eicosatetraenoic acid (EPA) and arachidonic acid (ARA). The first two compose the group known as Omega-3 and the last is Omega-6. While ARA is most often found in coastal benthic areas, DHA is preserved throughout the food web, increasing its contents with trophic position. Various authors have reported65,66 correlation with δ15N values,23,90 with results indicating δ15N values can be properly applied to determine the structure of food webs. It should be noted that although using multiple tracers (isotopes, Hg and fatty acids) is informative, the cost-benefit ratio of the analyzes should be considered. The strategy of grouping data previously collected for other purposes, such as nutritional studies (fatty acids) and investigations on contamination (Hg), can be more advantageous. Moreover, the tools have a strong potential for application at the individual level, such as in studies of populations of specific migratory species.23,89 Mixing and isotopic niche models Mixing models have been developed as more precise tools to investigate the relative contribution of food items to consumers91-93 and significant progress in their capabilities and sophistication has been achieved.25,94 Initial linear mass balance mixing models were restricted to systems involving a single consumer (or the average of multiple consumers). In addition, the number of sources permitted was limited by the number of isotopes utilized plus 1, which generally restricted the number of sources of the model to 3 (e.g., δ13C and δ15N + 1 = 3).7,95 Another limiting aspect is that these initial linear models did not provide measurements of errors or confidence intervals, which restricted the results to a range of possible contribution values of each source. One first improvement was the development of calculations of error propagation of each system through the IsoError model by Phillips and Gregg,92 which establishes confidence intervals around the estimates based on the variances of the consumer and the isotopic values of the sources. Later, the IsoSource model emerged as a linear solution to deal with many sources.92 The results are exhibited as frequency histograms, depicting all the viable contributions and descriptive statistics of the distributions of each food source. The model was further improved and simplified by Phillips et al.93 suggesting two ways to group sources that are isotopically and ecologically similar. However, the model continues to have limitations regarding the variability of the parameters and the investigation of ecological questions. Other models were developed shortly after the IsoSource, using different algorithms, but have been applied less often.7,25 Another question involving mixing models is that they do not consider the differences in the elemental concentrations of the resources eaten, instead assuming they are equal.91 Although this assumption is valid for some herbivores and carnivores, it often does not apply to omnivorous and generalist animals, since they feed on a wide variety of resources with different trophic levels. For this reason, the IsoConc model was developed to perform "concentration-dependent" calculations, including data on the elemental composition of sources, also considering digestibility.91 This was a significant advance, increasing the precision of estimates on the organisms' diet. However, its practical application to generalist animals is not that simple, and unlike IsoError, it does not allow including the errors associated with the process.7,25 In response to these limitations of standard linear models, Bayesian statistics have been incorporated in mixing models, generating significant advances in trophic approaches.25 The first model developed in this respect was MixSIR,96 which was soon followed by the SIAR26 and MixSIAR97 models. Although some characteristics have been improved (e.g., algorithms, graphical outputs), the essence of these models is similar. Both integrate the probability calculations regarding the relative contributions in systems with various sources, providing credibility intervals. These calculations perform corrections by including the isotopic fractionation, along with the associated standard deviation. Besides these models, Hopkins and Ferguson98 created the IsotopeR model, which integrates all the resources of the models previously described using entirely Bayesian calculations, including measurement of errors associated with isotopic analyzes, concentration dependence, isotopic correlation, and estimates at the individual level. Among the models mentioned, the SIAR has often been applied to investigate food assimilation of the organisms of interest.25,99 Figure 3 shows a graphical example of the SIAR used to examine the assimilation of prey items in the diet of two fish species. Despite all the advances from incorporating Bayesian statistics in trophic models, there are still challenges to their practical application. Phillips et al.25 made some recommendations for more effective use of mixing models in trophic ecology studies. First, these studies should involve clear questions, gather information about the functioning of the ecosystem, and have an experimental design that effectively encompasses the isotopic variability of consumers and their sources on appropriate temporal and spatial scales (considering, for instance, the turnover rate). In addition, researchers need to adequately choose the models applied according to the questions to be answered, recognizing their premises and limitations. The authors further stressed that the decisions about grouping or not grouping similar sources or including the dependence of concentration in the calculations could influence the results.
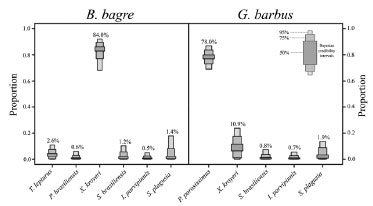 Figure 3. Example of the SIAR mixing model results on the contribution (%) of different prey items in the feeding habits of two fish species (Bagre bagre and Genidens barbus) (extracted and modified from Di Beneditto et al.32 and reprinted with Criative Commons permission CC-BY 4.0). X. kroyeri: Xiphopenaeus kroyeri (omnivore crustacean), I. parvipinnis: Isopisthus parvipinnis (carnivore fish), P. brasiliensis: Paralonchurus brasiliensis (benthic feeder fish), P. porosissimus: Porichthys porosissimus (carnivore fish), S. brasiliensis: Stellifer brasiliensis (benthic feeder fish), S. plagusia: Symphurus plagusia (benthic feeder fish), T. lepturus: Trichiurus lepturus (voracious predator fish)
In 2018, Healy et al.100 developed the SIDER Bayesian model to facilitate TEF coupled with mixing models. It consists of a phylogenetic regression model based on a dataset compiled to impute a TEF of a given consumer. The regression model is fitted by Bayesian inference and returns a posterior distribution describing the estimated TEF, which, in turn, can be used to calculate the mean value and its uncertainty (variance or standard deviation). These distributions are also compatible with all main Bayesian stable isotope mixing models, including MixSIAR;97 IsotopeR;98 SIAR;26 and MixSIR.96 As an advantage, SIDER allows estimating the TEF for species not yet included in the database, based on phylogenetically close species. However, the disadvantage, so far, is that SIDER has only terrestrial and marine birds and mammals in the database. It is noteworthy that SIDER was developed as an alternative solution when researchers have no TEF for its target species but phylogenetically close species to estimate it. However, experimental measurement with controlled feeding trials is the most efficient way to assess TEF for organisms.100,101 The isotopic niche and delta space of species or groups of species partly represents their ecological niches in the broad sense.102,103 Based on this, the concept of "isotopic niche" emerged, enabling its application for various purposes and areas of ecology.7,31,32,104 Many advances have also been achieved based on exploring the space formed by the dispersion of the set of isotopic data, enabling inferences about the isotopic niche and various aspects of trophic ecology. Traditionally, estimates of the niche amplitude were carried out by analyzing the stomach content and applying measures of richness and uniformity to groups of individuals. However, because of some limitations of these approaches, the use of isotope metrics has become a robust complement in the investigation of trophic niches.34,40,105,106 In this sense, Layman et al.24 formulated quantitative metrics generated from the dispersion of the isotope values to supply information about the niche amplitude and food web structure. In simplified form, the metrics reflect the trophic diversity according to the distribution of δ15N and δ13C values in a bi-dimensional space, indicating how individuals of a species are closely related within their respective niches. Estimates are generated on the variety of food items exploited, the trophic diversity, amplitude, redundancy (similarity), and uniformity of the distribution of individuals in the isotopic niche space. An important point to be considered is that the metrics are based on the total area of the polygon to estimate the trophic niche, covering all the dispersed values in the space of δ15N and δ13C values, including outliers. Nevertheless, when comparing niches with different sample sizes, this approach may not be the best suitable, since the area of the polygon tends to increase with the number of samples. In response, Jackson et al.28 suggested an adaptation of the metrics of Layman et al.,24 creating a proposal based on Bayesian inference called SIBER ("Stable Isotope Bayesian Ellipses in R"). The SIBER model uses the isotopic area of a standard ellipse (SEA, standard ellipse area). The standard ellipse is based on the variance and covariance of the isotope values, covering 40% of the data, representing the core of the isotopic niche. Hence, when these ellipses overlap, the compared groups share the same isotopic space and consequently can be used as a measure of the niche overlap.28,107 For this, the authors recommend a sample number of around 30. However, when working with smaller values, a correction factor can be applied in the calculation to adjust the SEA, which is called the corrected standard ellipse area (SEAc).28 By considering the variability existing in the process, this metric is better for comparison between spatial and temporal gradients, since the extreme values no longer have a significant effect on the measure. Besides this, the metrics are not affected by the errors associated with the sample number, allowing the comparison of groups with different sizes. Therefore, the approach identifies general patterns between other systems and sampling periods.28,108 For example, this permits comparing species within the same community (Figure 4A), in different communities (Figures 4B and 4C) and different sampling periods or seasons (Figure 4C).
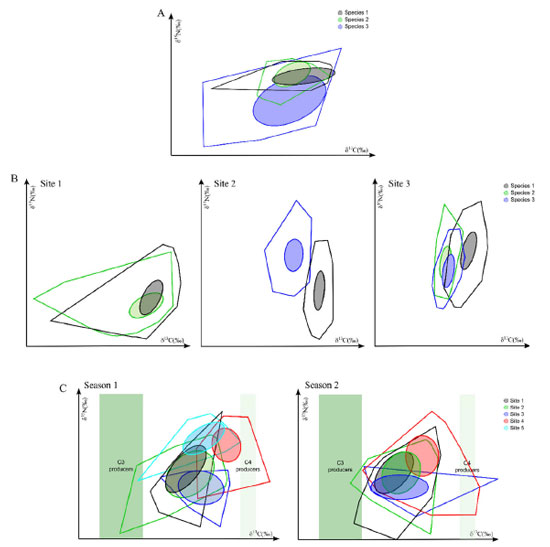 Figure 4. Graphical examples of studies using SIBER to investigate the isotopic niche in different situations. (A): Different catfish species within a community (modified from Gatts et al.);109 (B): comparing species of shrimps in different areas (modified from Ferreira et al.);110 and (C): comparing fish species between different areas and seasonal periods (modified from Abrantes et al.)108
The metrics generated by the Bayesian approach enable robust comparisons between communities, which is one of the strong points of this technique. Nevertheless, some caveats about its application should be considered. For example, when applied to entire communities using averages of small samples, the uncertainty increases. Therefore, even when using corrected SEA, authors recommend a minimum of 10 samples per group to be analyzed to mitigate these effects.28 It also should be stressed that the isotopic niche estimates should not be considered a direct measure of species' ecological niche. It should be interpreted as a marker that permits inferences about fundamental aspects of the ecological niche of species or communities.7,25,28 Table 1 summarizes the main models and approaches described and utilized in trophic studies, highlighting their advances, strong points, and limitations. Besides these models, two other Bayesian models can be mentioned, although their application in trophic studies has not been widely explored in the literature, namely the IsoWeb111 and FRUITS.112
Finally, the rapid and extensive growth of stable isotope data across diverse scientific fields, such as plant and animal physiology, paleobiology, evolution, biomedicine, climatology, and community and ecosystem ecology has prompted researchers from around the world to establish centralized databases to manage isotopic data.118 Recently, three such databases that cover aquatic habitats have been created: IsoBank,118 MarTurtSI,119 and SIA-BRA.120 IsoBank and MarTurtSI are international databases with isotopic data from various environmental and biological matrices and marine turtles, respectively, while SIA-BRA is a database developed by Brazilian scientists that includes data from terrestrial and aquatic animals found in Brazilian biomes and coastal marine areas. SIA-BRA contains approximately 44% of the data from coastal marine systems (oceanic plus estuarine). These databases are essential for advancing isotopic ecology, enabling studies on a range of topics such as diet tracing, foraging ecology, habitat use, food webs, effects of phylogeny on dietary ecology, and physiological studies.120 For aquatic food webs, isotopic databases are particularly important since they provide data to researchers who may not be able to sample or access all components of the food web.
FINAL CONSIDERATIONS In this literature review, we have presented the main approaches applied in the analysis of trophic ecology using stable isotopes, which can generate precise information to shed light on the dynamics of ecological communities. Application of the techniques summarized here can enable: (i) tracing the sources of organic matter in trophic chains; (ii) determining the contribution of each food source in the diet of organisms; (iii) investigating the vertical structure of food chains; (iv) making inferences about the use of resources and habitats of the species studied, generating estimates of niche amplitude; and (v) making robust estimates and comparisons through current statistical models. Researchers should bear in mind that all tools have premises, advantages, and drawbacks, which should be well understood for effective interpretation of the results. The application of multiple methods and understanding the life history of the target organisms and interactions among species will provide a better understanding of food webs. Besides this, the information generated by these tools is often complementary, so the application of multiple models can be a more appropriate alternative to overcome the limitations of each model and expand knowledge of aquatic food webs.
ACKNOWLEDGMENTS We are grateful to the Environmental Sciences Laboratory and the Graduate Program in Ecology and Natural Resources of Universidade Estadual do Norte Fluminense Darcy Ribeiro. We also acknowledge the financial support of the National Council for Scientific and Technological (CNPq - grants 305.217/2017-8 to C. E. R., and 302.598/2021-9 to A. P. M. D. B.) and Rio de Janeiro State Research Foundation (FAPERJ - grants E-26/210.703/2021 and E-26/200.893/2021 to C. R. R., E-26/200.797/2021 to A. P. M. D. B., and E-26/204.508/2021 to P. V. G.). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - finance code 001 (fellowship to C. P. F.).
REFERENCES 1. Odum, E. P.; Fundamentos de Ecologia, 6ª ed.; Fundação Calouste Gulbenkian: Lisboa, 2001. 2. Thompson, R. M.; Brose, U.; Dunne, J. A.; Hall, R. O.; Hladyz, S.; Kitching, R. L.; Martinez, N. D.; Rantala, H.; Romanuk, T. N.; Stouffer, D. B.; Tylianakis, J. M.; Trends Ecol. Evol. 2012, 27, 689. [Crossref] 3. Layman, C. A.; Giery, S. T.; Buhler, S.; Rossi, R.; Penland, T.; Henson, M. N.; Bogdanoff, A. K.; Cove, M. V.; Irizarry, A. D.; Schalk, C. M.; Archer, S. K.; Food Webs 2015, 4, 14. [Crossref] 4. Elton, C. In Animal Ecology; Huxley, J. S.; Hogben, L. T.; de Beer, C. R.; Garstang, W., eds.; The MacMillan Company: New York, 1927. 5. Lindeman, R. L.; Ecology 1942, 23, 399. [Crossref] 6. Vander Zanden, M. J.; Cabana, G.; Rasmussen, J. B.; Can. J. Fish. Aquat. Sci. 1997, 54, 1142. [Crossref] 7. Layman, C. A.; Araujo, M. S.; Boucek, R.; Hammerschlag-Peyer, C. M.; Harrison, E.; Jud, Z. R.; Matich, P.; Rosenblatt, A. E.; Vaudo, J. J.; Yeager, L. A.; Post, D. M.; Bearhop, S.; Biol. Rev. 2012, 87, 545. [Crossref] 8. DeNiro, M. J.; Epstein, S.; Geochim. Cosmochim. Acta 1978, 42, 495. [Crossref] 9. Fry, B.; Stable Isotope Ecology, 1st ed.; Springer: New York, 2006. 10. Martinelli, L. A.; de Camargo, P. B.; Ometto, J. P. H.; Ferraz, E. S.; Victoria, R. L.; Moreira, M. Z.; Desvendando Questões Ambientais com Isótopos Estáveis, 1ª ed.; Oficina de Textos: São Paulo, 2009. 11. dos Santos, V. H. J. M.; Rodrigues, L. F.; J. Braz. Chem. Soc. 2021, 32, 59. [Crossref] 12. Lima, B. D.; Martins, L. L.; Santos, L. C.; de Souza, E. S.; Pudenzi, M. A.; Nascimento, H. L.; Eberlin, M. N.; da Cruz, G. F.; J. Braz. Chem. Soc. 2020, 31, 673. [Crossref] 13. Leal, O. A.; Dick, D. P.; Costa, F. S.; Knicker, H.; de Carvalho Júnior, J. A.; Santos, J. C.; J. Braz. Chem. Soc. 2019, 30, 413. [Crossref] 14. Ceccopieri, M.; Scofield, A. L.; Almeida, L.; Wagener, A. L. R.; J. Braz. Chem. Soc. 2018, 29, 2363. [Crossref] 15. Post, D. M.; Ecology 2002, 83, 703. [Crossref] 16. Carr, M. K.; Jardine, T. D.; Doig, L. E.; Jones, P. D.; Bharadwaj, L.; Tendler, B.; Chételat, J.; Cott, P.; Lindenschmidt, K. E.; Sci. Total Environ. 2017, 586, 338. [Crossref] 17. Willacker, J. J.; Eagles-Smith, C. A.; Ackerman, J. T.; Environ. Sci. Technol. 2017, 51, 2131. [Crossref] 18. Pizzochero, A. C.; Michel, L. N.; Chenery, S. R.; McCarthy, I. D.; Vianna, M.; Malm, O.; Lepoint, G.; Das, K.; Dorneles, P. R.; Can. J. Fish. Aquat. Sci. 2018, 75, 977. [Crossref] 19. Di Beneditto, A. P. M.; Bittar, V. T.; Camargo, P. B.; Rezende, C. E.; Kehrig, H. A.; Arch. Environ. Contam. Toxicol. 2012, 62, 264. [Crossref] 20. Kehrig, H. A.; Baptista, G.; Di Beneditto, A. P. M.; Almeida, M. G.; Rezende, C. E.; Siciliano, S.; De Moura, J. F.; Moreira, I.; Revista de Biología Marina y Oceanografía 2017, 52, 233. [Crossref] 21. Nielsen, J. M.; Popp, B. N.; Winder, M.; Oecologia 2015, 178, 631. [Crossref] 22. Brett, M. T.; Eisenlord, M. E.; Galloway, A. W. E.; Ecosphere 2016, 7, e01440. [Crossref] 23. Sardenne, F.; Hollanda, S.; Lawrence, S.; Albert-Arrisol, R.; Degroote, M.; Bodin, N.; Ecol. Indic. 2017, 81, 315. [Crossref] 24. Layman, C. A.; Arrington, D. A.; Montaña, C. G.; Post, D. M.; Ecology 2007, 88, 42. [Link] accessed in October 2023 25. Phillips, D. L.; Inger, R.; Bearhop, S.; Jackson, A. L.; Moore, J. W.; Parnell, A. C.; Semmens, B. X.; Ward, E. J.; Can. J. Zool. 2014, 92, 823. [Crossref] 26. Parnell, A. C.; Inger, R.; Bearhop, S.; Jackson, A. L.; PLoS One 2010, 5, 1. [Crossref] 27. Bond, A. L.; Diamond, A. W.; Ecological Applications 2011, 21, 1017. [Crossref] 28. Jackson, A. L.; Inger, R.; Parnell, A. C.; Bearhop, S.; J. Anim. Ecol. 2011, 80, 595. [Crossref] 29. Fry, B.; Mar. Ecol.: Prog. Ser. 2013, 472, 1. [Crossref] 30. Abdurahiman, K. P.; Nayak, T. H.; Zacharia, P. U.; Mohamed, K. S.; Estuarine Coastal Shelf Sci. 2010, 87, 601. [Crossref] 31. Di Beneditto, A. P. M.; Monteiro, L. R.; J. Mar. Biol. Assoc. U. K. 2016, 96, 853. [Crossref] 32. Di Beneditto, A. P. M.; Tavares, M. T. M.; Monteiro, L. R.; Biota Neotropica 2018, 18, 1. [Crossref] 33. Philippsen, J. S.; Benedito, E.; Oecologia Australis 2013, 17, 15. [Crossref] 34. Bearhop, S.; Adams, C. E.; Waldron, S.; Fuller, R. A.; Macleod, H.; J. Anim. Ecol. 2004, 73, 1007. [Crossref] 35. Boecklen, W. J.; Yarnes, C. T.; Cook, B. A.; James, A. C.; Annual Review of Ecology, Evolution, and Systematics 2011, 42, 411. [Crossref] 36. Vander Zanden, M.; Rasmussen, J.; Limnol. Oceanogr. 2001, 46, 2061. [Crossref] 37. Lavoie, R. A.; Jardine, T. D.; Chumchal, M. M.; Kidd, K. A.; Campbell, L. M.; Environ. Sci. Technol. 2013, 47, 13385. [Crossref] 38. Hussey, N. E.; Macneil, M. A.; Mcmeans, B. C.; Olin, J. A.; Dudley, S. F. J.; Cliff, G.; Wintner, S. P.; Fennessy, S. T.; Fisk, A. T.; Ecology Letters 2014, 17, 239. [Crossref] 39. Ferreira, K. A.; Braga, A. A.; Di Beneditto, A. P. M.; Ocean & Coastal Management 2021, 205, 105500. [Crossref] 40. Di Beneditto, A. P. M.; Rezende, C. E.; Camargo, P. B.; Kehrig, H. A.; Biota Neotropica 2013, 13, 29. [Crossref] 41. Milmann, L.; Machado, R.; Sucunza, F.; de Oliveira, L. R.; dos Santos, R. A.; Di Beneditto, A. P. M.; Rezende, C. E.; Baumgarten, J.; Ott, P. H.; Mammalia 2018, 83, 49. [Crossref] 42. Agostinho, K. F. F.; Monteiro, L. R.; Di Beneditto, A. P. M.; Biota Neotropica 2021, 21, e20201099. [Crossref] 43. Gatts, P. V.; Franco, M. A. L.; Almeida, M. G.; Rezende, C. E.; Costa, P. A. S.; Estuarine Coastal Shelf Sci. 2021, 261, 107563. [Crossref] 44. Connolly, R. M.; Guest, M. A.; Melville, A. J.; Oakes, J. M.; Oecologia 2004, 138, 161. [Crossref] 45. Clayden, M. G.; Lescord, G. L.; Kidd, K. A.; Wang, X.; Muir, D. C. G.; O'Driscoll, N. J.; Environ. Toxicol. Chem. 2017, 36, 661. [Crossref] 46. McCutchan, J. H.; Lewis, W. M.; McGrath, C. C.; Oikos 2003, 102, 378. [Crossref] 47. Hobson, K. A.; Clark, R. G.; Condor 1992, 94, 181. [Crossref] 48. Manetta, G. I.; Cecilio, E. B.; Acta Sci., Biol. Sci. 2003, 25, 121. [Crossref] 49. Barnes, C.; Jennings, S.; Rapid Commun. Mass Spectrom. 2007, 21, 1461. [Crossref] 50. Florin, S. T.; Felicetti, L. A.; Robbins, C. T.; Funct. Ecol. 2011, 25, 519. [Crossref] 51. Caut, S.; Angulo, E.; Courchamp, F.; J. Appl. Ecol. 2009, 46, 443. [Crossref] 52. Caut, S.; Angulo, E.; Courchamp, F.; Figuerola, J.; J. Appl. Ecol. 2010, 47, 948. [Crossref] 53. Wang, X.; Bao, Z.; Lin, C. J.; Yuan, W.; Feng, X.; Environ. Sci. Technol. 2016, 50, 8548. [Crossref] 54. Liu, G.; Cai, Y.; O'Driscoll, N.; Environmental Chemistry and Toxicology of Mercury, 1st ed.; John Wiley and Sons: New Jersey, 2011. 55. Araujo, B. F.; Almeida, M. G.; Salomão, M. S. M. B.; Gobo, R. R.; Siqueira, V. C.; Ovalle, A. R.; Rezende, C. E.; Quim. Nova 2010, 33, 501. [Crossref] 56. Correia, R. R. S.; Guimarães, J. R. D.; Chemosphere 2017, 167, 438. [Crossref] 57. Kehrig, H. A.; Palermo, E. F. A.; Seixas, T. G.; Santos, H. S. B.; Malm, O.; Akagi, H.; J. Braz. Chem. Soc. 2009, 20, 1142. [Crossref] 58. Kehrig, H. A.; Costa, M.; Moreira, I.; Malm, O.; J. Braz. Chem. Soc. 2006, 17, 1409. [Crossref] 59. Kehrig, H. A.; Malm, O.; Palermo, E. F. A.; Seixas, T. G.; Baêta, A. P.; Moreira, I.; Quim. Nova 2011, 34, 377. [Crossref] 60. Rosera, T. J.; Janssen, S. E.; Tate, M. T.; Lepak, R. F.; Ogorek, J. M.; DeWild, J. F.; Krabbenhoft, D. P.; Hurley, J. P.; ACS ES&T Water 2022, 2, 701. [Crossref] 61. Ullrich, S. M.; Tanton, T. W.; Abdrashitova, S. A.; Crit. Rev. Environ. Sci. Technol. 2001, 31, 241. [Crossref] 62. Chen, C.; Amirbahman, A.; Fisher, N.; Harding, G.; Lamborg, C.; Nacci, D.; Taylor, D.; EcoHealth 2008, 5, 399. [Crossref] 63. Seixas, T. G.; Moreira, I.; Malm, O.; Kehrig, H. A.; J. Braz. Chem. Soc. 2012, 23, 1280. [Crossref] 64. Lima, G.; Menegario, A.; Sulato, E.; Pedrobom, J.; Torres-Florez, J.; de Araújo Júnior, M.; Barreto, A.; J. Braz. Chem. Soc. 2022, 33, 1309. [Crossref] 65. Kehrig, H. A.; Fernandes, K. W. G.; Malm, O.; Seixas, T. G.; Di Beneditto, A. P. M.; Souza, C. M. M.; Quim. Nova 2009, 32, 1822. [Crossref] 66. Tovar, L. R.; Neves, M. C.; Manhães, B. M. R.; Montanini, G.; Azevedo, A. F.; Lailson-Brito, J.; Bisi, T. L.; Chemosphere 2023, 338, 139496. [Crossref] 67. Di Beneditto, A. P. M.; Bittar, V. T.; de Rezende, C. E.; Camargo, P. B.; Kehrig, H. A.; Neotropical Ichthyology 2013, 11, 211. [Crossref] 68. Kehrig, H. A.; Seixas, T. G.; Malm, O.; Di Beneditto, A. P. M.; Rezende, C. E.; Mar. Pollut. Bull. 2013, 75, 283. [Crossref] 69. Jones, H. J.; Swadling, K. M.; Butler, E. C. V.; Barry, L. A.; Macleod, C. K.; Limnol. Oceanogr. 2014, 59, 1181. [Crossref] 70. Baptista, G.; Kehrig, H. A.; Di Beneditto, A. P. M.; Hauser-Davis, R. A.; Almeida, M. G.; Rezende, C. E.; Siciliano, S.; de Moura, J. F.; Moreira, I.; Environ. Pollut. 2016, 218, 1298. [Crossref] 71. Di Beneditto, A. P. M.; Kehrig, H. A.; Pestana, I. A.; Bull. Environ. Contam. Toxicol. 2021, 107, 124. [Crossref] 72. Blum, J. D.; Sherman, L. S.; Johnson, M. W.; Annu. Rev. Earth Planet. Sci. 2014, 42, 249. [Crossref] 73. Eckley, C. S.; Gilmour, C. C.; Janssen, S.; Luxton, T. P.; Randall, P. M.; Whalin, L.; Austin, C.; Sci. Total Environ. 2020, 707, 136031. [Crossref] 74. Tsui, M. T. K.; Blum, J. D.; Kwon, S. Y.; Sci. Total Environ. 2020, 716, 135386. [Crossref] 75. Bartrons, M.; Gratton, C.; Spiesman, B. J.; Zanden, M. J. V.; Ecological Applications 2015, 25, 151. [Crossref] 76. Fry, B.; Chumchal, M. M.; Ecological Applications 2012, 22, 606. [Crossref] 77. Araujo, B. F.; Hintelmann, H.; Dimock, B.; Almeida, M. G.; Rezende, C. E.; Chemosphere 2017, 178, 42. [Crossref] 78. Perrot, V.; Landing, W. M.; Grubbs, R. D.; Salters, V. J. M.; Sci. Total Environ. 2019, 666, 828. [Crossref] 79. Yuan, J.; Liu, Y.; Chen, S.; Peng, X.; Li, Y. F.; Li, S.; Zhang, R.; Zheng, W.; Chen, J.; Sun, R.; Heimbürger-Boavida, L. E.; Environ. Sci. Technol. 2023, 57, 6550. [Crossref] 80. Chikaraishi, Y.; Steffan, S. A.; Ogawa, N. O.; Ishikawa, N. F.; Sasaki, Y.; Tsuchiya, M.; Ohkouchi, N.; Ecology and Evolution 2014, 4, 2423. [Crossref] 81. Thorp, J. H.; Bowes, R. E.; Ecosystems 2017, 20, 1029. [Crossref] 82. Bowes, R. E.; Thorp, J. H.; Ecosphere 2015, 6, 1. [Crossref] 83. McMahon, K. W.; Thorrold, S. R.; Elsdon, T. S.; McCarthy, M. D.; Limnol. Oceanogr. 2015, 60, 1076. [Crossref] 84. Chikaraishi, Y.; Ogawa, N. O.; Kashiyama, Y.; Takano, Y.; Suga, H.; Tomitani, A.; Miyashita, H.; Kitazato, H.; Ohkouchi, N.; Limnol. Oceanogr.: Methods 2009, 7, 740. [Crossref] 85. Blanke, C. M.; Chikaraishi, Y.; Takizawa, Y.; Steffan, S. A.; Dharampal, P. S.; Vander Zanden, M. J.; Can. J. Fish. Aquat. Sci. 2017, 74, 1291. [Crossref] 86. Dalsgaard, J.; St. John, M.; Kattner, G.; Müller-Navarra, D.; Hagen, W.; Adv. Mar. Biol. 2003, 46, 225. [Crossref] 87. Iverson, S. J.; Field, C.; Don Bowen, W.; Blanchard, W.; Ecol. Monogr. 2004, 74, 211. [Crossref] 88. Hall, D.; Lee, S. Y.; Meziane, T.; J. Exp. Mar. Biol. Ecol. 2006, 336, 42. [Crossref] 89. Brewster, J. D.: Characterizing the Diet and Habitat Niches of Coastal Fish Populations in the Beaufort Sea Tarium Niryutait Marine Protected Area; MSc dissertation, University of Manitoba, Canada, 2016. [Link] accessed in October 2023 90. Koussoroplis, A. M.; Bec, A.; Perga, M. E.; Koutrakis, E.; Bourdier, G.; Desvilettes, C.; Estuarine Coastal Shelf Sci. 2011, 91, 450. [Crossref] 91. Phillips, D. L.; Koch, P. L.; Oecologia 2002, 130, 114. [Crossref] 92. Phillips, D. L.; Gregg, J. W.; Oecologia 2003, 136, 261. [Crossref] 93. Phillips, D. L.; Newsome, S. D.; Gregg, J. W.; Oecologia 2005, 144, 520. [Crossref] 94. Shipley, O. N.; Matich, P.; Oecologia 2020, 193, 27. [Crossref] 95. Phillips, D. L.; Oecologia 2001, 127, 166. [Crossref] 96. Moore, J. W.; Semmens, B. X.; Ecology Letters 2008, 11, 470. [Crossref] 97. Stock, B.; Jackson, A.; Ward, E.; Venkiteswaran, J.; MixSIAR, version 3.1; R Core Team, Austria, 2016. [Crossref] 98. Hopkins, J. B.; Ferguson, J. M.; PLoS One 2012, 7, e28478. [Crossref] 99. Parnell, A. C.; Phillips, D. L.; Bearhop, S.; Semmens, B. X.; Ward, E. J.; Moore, J. W.; Jackson, A. L.; Grey, J.; Kelly, D. J.; Inger, R.; Environmetrics 2013, 24, 387. [Crossref] 100. Healy, K.; Guillerme, T.; Kelly, S. B. A.; Inger, R.; Bearhop, S.; Jackson, A. L.; Ecography 2018, 41, 1393. [Crossref] 101. Del Rio, C. M.; Wolf, N.; Carleton, S. A.; Gannes, L. Z.; Biol. Rev. 2009, 84, 91. [Crossref] 102. Newsome, S. D.; Del Rio, C.; Bearhop, S.; Phillips, D. L.; Front. Ecol. Environ. 2007, 5, 429. [Crossref] 103. Newsome, S. D.; Yeakel, J. D.; Wheatley, P. V.; Tinker, M. T.; J. Mammal. 2012, 93, 329. [Crossref] 104. Jackson, M. C.; Donohue, I.; Jackson, A. L.; Britton, J. R.; Harper, D. M.; Grey, J.; PLoS One 2012, 7, 1. [Crossref] 105. Di Beneditto, A. P. M.; de Souza, C. M. M.; Kehrig, H. A.; Rezende, C. E.; Mar. Biol. 2011, 158, 2209. [Crossref] 106. Costa, P. A. S.; Braga, A. C.; Malavolti, G. S.; Franco, M. A. L.; Gatts, P. V.; Batista, A.; Rezende, C. E.; J. Mar. Biol. Assoc. U. K. 2019, 99, 1399. [Crossref] 107. Knickle, D. C.; Rose, G. A.; Environ. Biol. Fishes 2014, 97, 343. [Crossref] 108. Abrantes, K. G.; Barnett, A.; Bouillon, S.; Funct. Ecol. 2014, 28, 270. [Crossref] 109. Gatts, P. V.; Franco, M. A. L.; Almeida, M. G.; Zalmon, I. R.; Di Beneditto, A. P. M.; Costa, P. A. S.; de Rezende, C. E.; J. Mar. Biol. Assoc. U. K. 2020, 100, 133. [Crossref] 110. Ferreira, K. A.; Braga, A. A.; Di Beneditto, A. P. M.; J. Mar. Biol. Assoc. U. K. 2022, 102, 338. [Crossref] 111. Kadoya, T.; Osada, Y.; Takimoto, G.; PLoS One 2012, 7, e41057. [Crossref] 112. Fernandes, R.; Millard, A. R.; Brabec, M.; Nadeau, M. J.; Grootes, P.; PLoS One 2014, 9, e87436. [Crossref] 113. Kline Junior, T. C.; Goering, J. J.; Mathisen, O. A.; Poe, P. H.; Parker, P. L.; Scalan, R. S.; Can. J. Fish. Aquat. Sci. 1993, 50, 2350. [Crossref] 114. Ben-David, M.; Flynn, R. W.; Schell, D. M.; Oecologia 1997, 111, 280. [Crossref] 115. Whitledge, G. W.; Rabeni, C. F.; Can. J. Fish. Aquat. Sci. 1997, 54, 2555. [Crossref] 116. Schwarcz, H. P.; J. Archaeol. Sci. 1991, 18, 261. [Crossref] 117. Phillips, D. L.; Gregg, J. W.; Oecologia 2001, 127, 171. [Crossref] 118. Pauli, J. N.; Newsome, S. D.; Cook, J. A.; Harrod, C.; Steffan, S. A.; Baker, C. J. O.; Ben-David, M.; Bloom, D.; Bowen, G. J.; Cerling, T. E.; Cicero, C.; Cook, C.; Dohm, M.; Dharampal, P. S.; Graves, G.; Gropp, R.; Hobson, K. A.; Jordan, C.; MacFadden, B.; Pilaar Birch, S.; Poelen, J.; Ratnasingham, S.; Russell, L.; Stricker, C. A.; Uhen, M. D.; Yarnes, C. T.; Hayden, B.; Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 2997. [Crossref] 119. Figgener, C.; Bernardo, J.; Plotkin, P. T.; Sci. Data 2019, 6, 1. [Crossref] 120. Diniz‐Reis, T. R.; Augusto, F. G.; Abdalla Filho, A. L.; Araújo, M. G. S.; Chaves, S. S. F.; Almeida, R. F.; Perez, E. B.; Simon, C. P.; Souza, J. L.; Costa, C. F. G.; Gomes, T. F.; Martinez, M. G.; Soltangheisi, A.; Mariano, E.; Vanin, A. S.; de Andrade, T. R.; Boesing, A. L.; Costa, F. J. V.; Fortuna, M. D.; Guedes, V. M.; Kisaka, T. B.; Kruszynski, C.; Lara, N. R. F.; Lima, R. A. M.; Pompermaier, V. T.; Rangel, B. S.; Ribeiro, J. F.; Santi Junior, A.; Tassoni Filho, M.; Ferreira, A.; Marques, T. S.; Pereira, A. L.; Aguiar, L. M. S.; dos Anjos, M. B.; Medeiros, E. S. F.; Benedito, E.; Calheiros, D. F.; Christofoletti, R. A.; Cremer, M. J.; Duarte‐Neto, P. J.; Nardoto, G. B.; de Oliveira, A. C. B.; Rezende, C. E.; da Silva, M. N. F.; Zuanon, J. A. S.; Verdade, L. M.; Moreira, M. Z.; de Camargo, P. B.; Martinelli, L. A.; Global Ecology and Biogeography 2022, 31, 611. [Crossref] |
On-line version ISSN 1678-7064 Printed version ISSN 0100-4042
Qu�mica Nova
Publica��es da Sociedade Brasileira de Qu�mica
Caixa Postal: 26037
05513-970 S�o Paulo - SP
Tel/Fax: +55.11.3032.2299/+55.11.3814.3602
Free access